Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Chen, X., Zhao, H., Wang, C., Hamed, M., Shang, Q., Yang, Y., Diao, X., Sun, X., Hu, W., Jiang, X., Zhang, Y., Hirsch, A.K.H., Wu, D., Zhuang, J.(2024) Int J Antimicrob Agents 63: 107160-107160
- PubMed: 38537721
- DOI: https://doi.org/10.1016/j.ijantimicag.2024.107160
- Primary Citation of Related Structures:
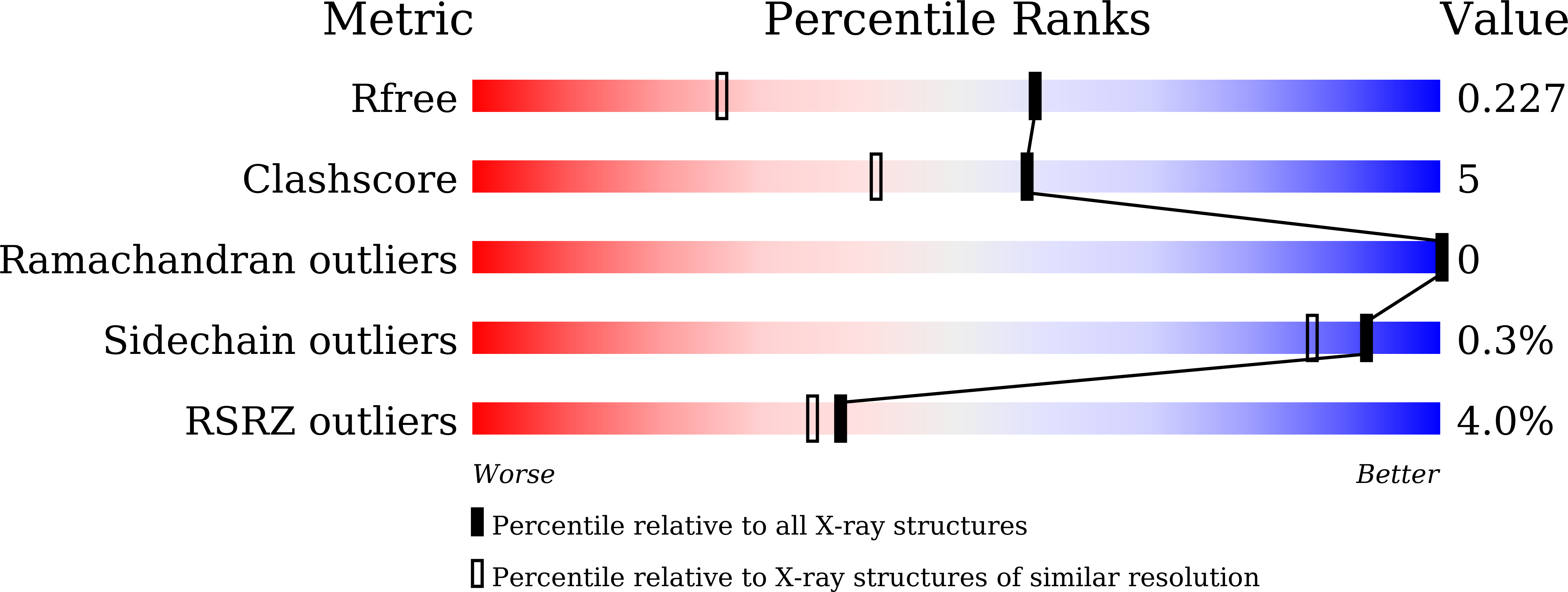
8XHU, 8XKF, 8XKG - PubMed Abstract:
In a vast majority of bacteria, protozoa and plants, the methylerythritol phosphate (MEP) pathway is utilized for the synthesis of isopentenyl diphosphate (IDP) and dimethylallyl diphosphate (DMADP), which are precursors for isoprenoids. Isoprenoids, such as cholesterol and coenzyme Q, play a variety of crucial roles in physiological activities, including cell-membrane formation, protein degradation, cell apoptosis, and transcription regulation. In contrast, humans employ the mevalonate (MVA) pathway for the production of IDP and DMADP, rendering proteins in the MEP pathway appealing targets for antimicrobial agents. This pathway consists of seven consecutive enzymatic reactions, of which 4-diphosphocytidyl-2C-methyl-D-erythritol synthase (IspD) and 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF) catalyze the third and fifth steps, respectively. In this study, we characterized the enzymatic activities and protein structures of Helicobacter pylori IspDF and Acinetobacter baumannii IspD. Then, using the direct interaction-based thermal shift assay, we conducted a compound screening of an approved drug library and identified 27 hit compounds potentially binding to AbIspD. Among them, two natural products, rosmarinic acid and tanshinone IIA sodium sulfonate, exhibited inhibitory activities against HpIspDF and AbIspD, by competing with one of the substrates, MEP. Moreover, tanshinone IIA sodium sulfonate also demonstrated certain antibacterial effects against H. pylori. In summary, we identified two IspD inhibitors from approved ingredients, broadening the scope for antibiotic discovery targeting the MEP pathway.
Organizational Affiliation:
Helmholtz International Lab, State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China.