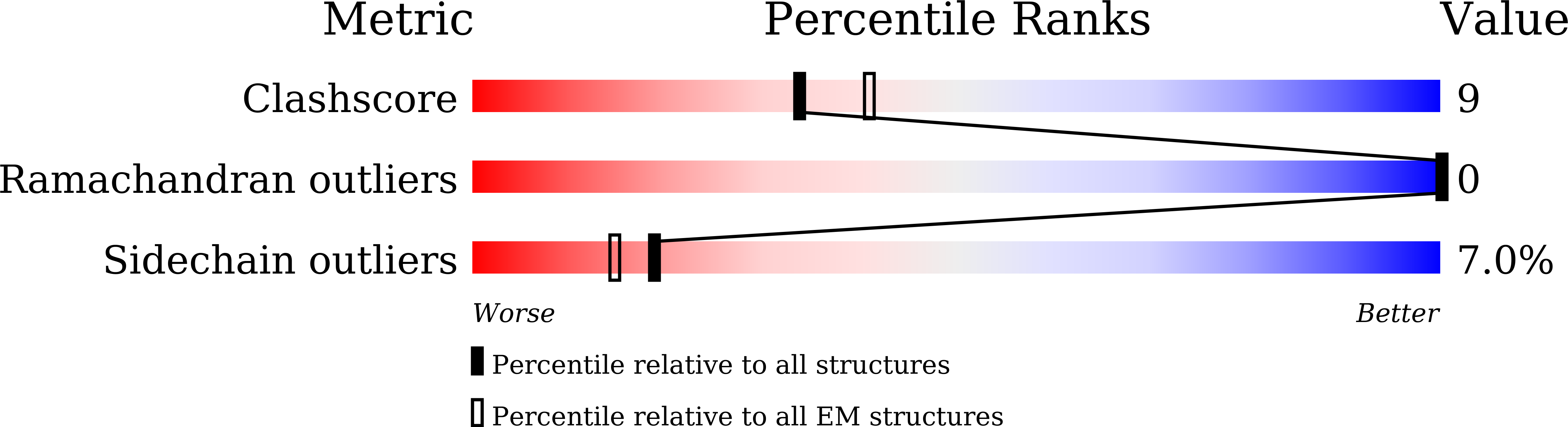Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Zhang, J.T., Liu, X.Y., Li, Z., Wei, X.Y., Song, X.Y., Cui, N., Zhong, J., Li, H., Jia, N.(2024) Nat Commun 15: 2797-2797
- PubMed: 38555355
- DOI: https://doi.org/10.1038/s41467-024-47177-9
- Primary Citation of Related Structures:
8WY8, 8WY9, 8WYA, 8WYB, 8WYC, 8WYD, 8WYE, 8WYF - PubMed Abstract:
Silent information regulator 2 (Sir2) proteins typically catalyze NAD + -dependent protein deacetylation. The recently identified bacterial Sir2 domain-containing protein, defense-associated sirtuin 2 (DSR2), recognizes the phage tail tube and depletes NAD + to abort phage propagation, which is counteracted by the phage-encoded DSR anti-defense 1 (DSAD1), but their molecular mechanisms remain unclear. Here, we determine cryo-EM structures of inactive DSR2 in its apo form, DSR2-DSAD1 and DSR2-DSAD1-NAD + , as well as active DSR2-tube and DSR2-tube-NAD + complexes. DSR2 forms a tetramer with its C-terminal sensor domains (CTDs) in two distinct conformations: CTD closed or CTD open . Monomeric, rather than oligomeric, tail tube proteins preferentially bind to CTD closed and activate Sir2 for NAD + hydrolysis. DSAD1 binding to CTD open allosterically inhibits tube binding and tube-mediated DSR2 activation. Our findings provide mechanistic insight into DSR2 assembly, tube-mediated DSR2 activation, and DSAD1-mediated inhibition and NAD + substrate catalysis in bacterial DSR2 anti-phage defense systems.
Organizational Affiliation:
Department of Biochemistry, School of Medicine, Southern University of Science and Technology, 518055, Shenzhen, China.