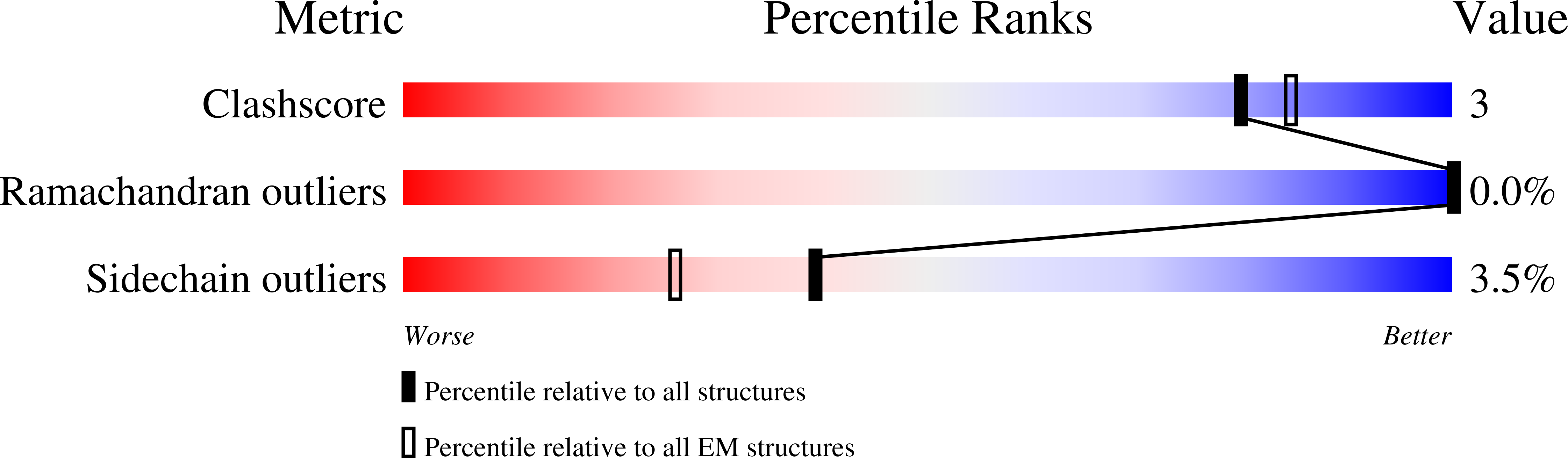Integrative structural analysis of Pseudomonas phage DEV reveals a genome ejection motor.
Cingolani, G., Lokareddy, R., Hou, C.F., Forti, F., Iglesias, S., Li, F., Pavlenok, M., Niederweis, M., Briani, F.(2024) Res Sq
- PubMed: 38463957
- DOI: https://doi.org/10.21203/rs.3.rs-3941185/v1
- Primary Citation of Related Structures:
8VXQ - PubMed Abstract:
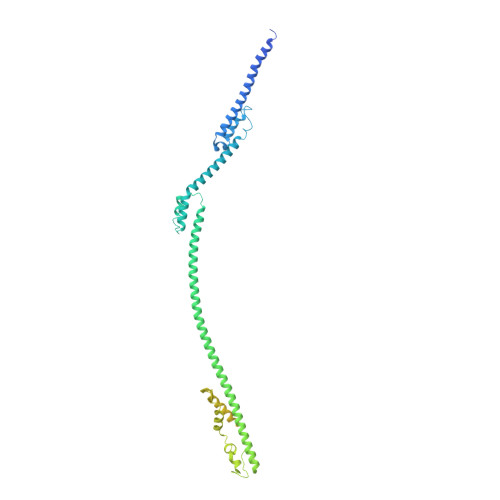
DEV is an obligatory lytic Pseudomonas phage of the N4-like genus, recently reclassified as Schitoviridae . The DEV genome encodes 91 ORFs, including a 3,398 amino acid virion-associated RNA polymerase. Here, we describe the complete architecture of DEV, determined using a combination of cryo-electron microscopy localized reconstruction, biochemical methods, and genetic knockouts. We built de novo structures of all capsid factors and tail components involved in host attachment. We demonstrate that DEV long tail fibers are essential for infection of Pseudomonas aeruginosa and dispensable for infecting mutants with a truncated lipopolysaccharide devoid of the O-antigen. We identified DEV ejection proteins and, unexpectedly, found that the giant DEV RNA polymerase, the hallmark of the Schitoviridae family, is an ejection protein. We propose that DEV ejection proteins form a genome ejection motor across the host cell envelope and that these structural principles are conserved in all Schitoviridae .
Organizational Affiliation:
University of Alabama at Birmingham.