Minimising the payload solvent exposed hydrophobic surface area optimises the antibody-drug conjugate properties.
Hobson, A.D., Zhu, H., Qiu, W., Judge, R.A., Longenecker, K.(2024) RSC Med Chem 15: 832-838
- PubMed: 38516584
- DOI: https://doi.org/10.1039/d3md00540b
- Primary Citation of Related Structures:
8VKZ - PubMed Abstract:
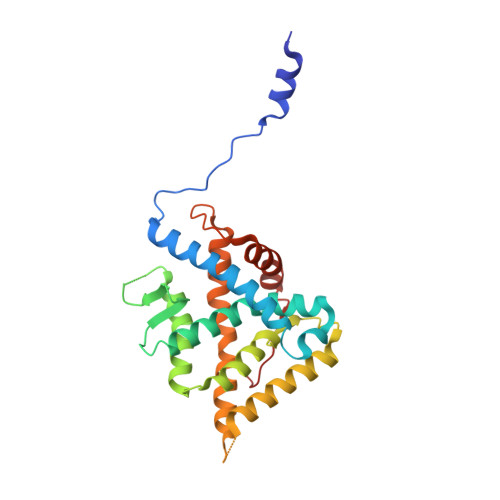
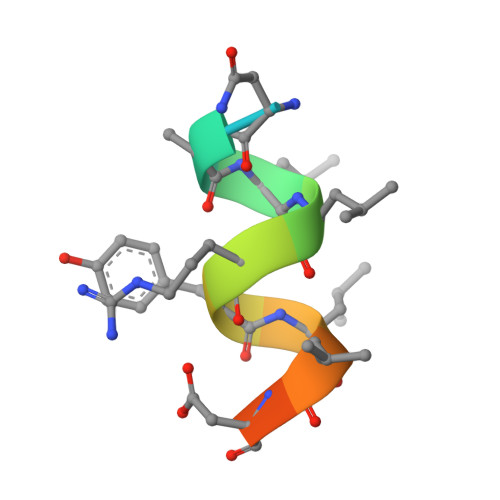
Glucocorticoid receptor modulators (GRMs) are an established and successful compound class for the treatment of multiple diseases. In addition, they are an area of high interest as payloads for antibody-drug conjugate s(ADCs) in both immunology and oncology. Solving the crystal structure of compound 2, the GRM payload from ABBV-3373 and ABBV-154, in the ligand binding domain of the glucocorticoid receptor (GR) revealed key information to facilitate optimal ADC payload design. All four critical H-bonds between the oxygen functional groups on the hexadecahydro-1 H -cyclopenta[ a ]phenanthrene ring system of the small molecule and protein were shown to be made (carbonyl at C3 to Gln570 and Arg611 and Asn564, carbonyl at C20 to Thr739, hydroxyl at C21 to Asn 564 and Thr739). In addition, an extra H-bond between the linker attachment site on compound 2, the aniline in the biaryl region, was observed. Confirmation of the stereochemistry of the acetal in compound 2 as ( R ) was established. Finally, the importance of minimising the exposed hydrophobic surface area of a payload to reduce the negative impact on the properties of resulting ADCs, like aggregation, was rationalised by comparison of ( R )-acetal compound 2 and ( S )-acetal compound 3.
Organizational Affiliation:
AbbVie Bioresearch Center 381 Plantation Street Worcester Massachusetts 01605 USA adrian.hobson@abbvie.com.