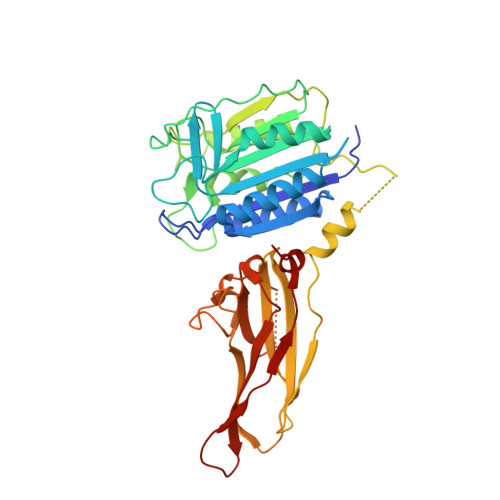
Inhibition of MALT1 and BCL2 induces synergistic anti-tumor activity in models of B cell lymphoma.
Plotnik, J.P., Richardson, A.E., Yang, H., Rojas, E., Bontcheva, V., Dowell, C., Parsons, S., Wilson, A., Ravanmehr, V., Will, C., Jung, P., Zhu, H., Partha, S.K., Panchal, S.C., Mali, R.S., Kohlhapp, F.J., McClure, R.A., Ramathal, C.Y., George, M.D., Jhala, M., Elsen, N.L., Qiu, W., Judge, R.A., Pan, C., Mastracchio, A., Henderson, J., Meulbroek, J.A., Green, M.R., Pappano, W.N.(2024) Mol Cancer Ther
- PubMed: 38507740
- DOI: https://doi.org/10.1158/1535-7163.MCT-23-0518
- Primary Citation of Related Structures:
8V4X - PubMed Abstract:
The activated B cell (ABC) subset of diffuse large B cell lymphoma (DLBCL) is characterized by chronic B cell receptor signaling and associated with poor outcomes when treated with standard therapy. In ABC-DLBCL, MALT1 is a core enzyme that is constitutively activated by stimulation of the B cell receptor or gain-of-function mutations in upstream components of the signaling pathway, making it an attractive therapeutic target. We discovered a novel small molecule inhibitor, ABBV-MALT1, that potently shuts down B cell signaling selectively in ABC-DLBCL preclinical models leading to potent cell growth and xenograft inhibition. We also identified a rational combination partner for ABBV-MALT1 in the BCL2 inhibitor, venetoclax, which when combined significantly synergizes to elicit deep and durable responses in preclinical models. This work highlights the potential of ABBV-MALT1 monotherapy and combination with venetoclax as effective treatment options for patients with ABC-DLBCL.
Organizational Affiliation:
AbbVie (United States), United States.