Structural and biochemical investigations of a HEAT-repeat protein involved in the cytosolic iron-sulfur cluster assembly pathway.
Vasquez, S., Marquez, M.D., Brignole, E.J., Vo, A., Kong, S., Park, C., Perlstein, D.L., Drennan, C.L.(2023) Commun Biol 6: 1276-1276
- PubMed: 38110506
- DOI: https://doi.org/10.1038/s42003-023-05579-3
- Primary Citation of Related Structures:
8USP, 8USQ - PubMed Abstract:
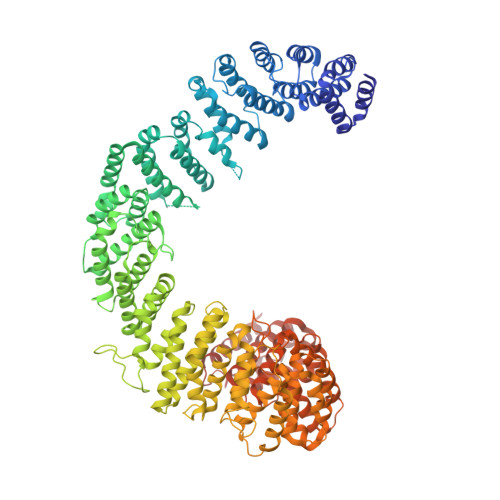
Iron-sulfur clusters are essential for life and defects in their biosynthesis lead to human diseases. The mechanism of cluster assembly and delivery to cytosolic and nuclear client proteins via the cytosolic iron-sulfur cluster assembly (CIA) pathway is not well understood. Here we report cryo-EM structures of the HEAT-repeat protein Met18 from Saccharomyces cerevisiae, a key component of the CIA targeting complex (CTC) that identifies cytosolic and nuclear client proteins and delivers a mature iron-sulfur cluster. We find that in the absence of other CTC proteins, Met18 adopts tetrameric and hexameric states. Using mass photometry and negative stain EM, we show that upon the addition of Cia2, these higher order oligomeric states of Met18 disassemble. We also use pulldown assays to identify residues of critical importance for Cia2 binding and recognition of the Leu1 client, many of which are buried when Met18 oligomerizes. Our structures show conformations of Met18 that have not been previously observed in any Met18 homolog, lending support to the idea that a highly flexible Met18 may be key to how the CTC is able to deliver iron-sulfur clusters to client proteins of various sizes and shapes, i.e. Met18 conforms to the dimensions needed.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, 02139, USA.