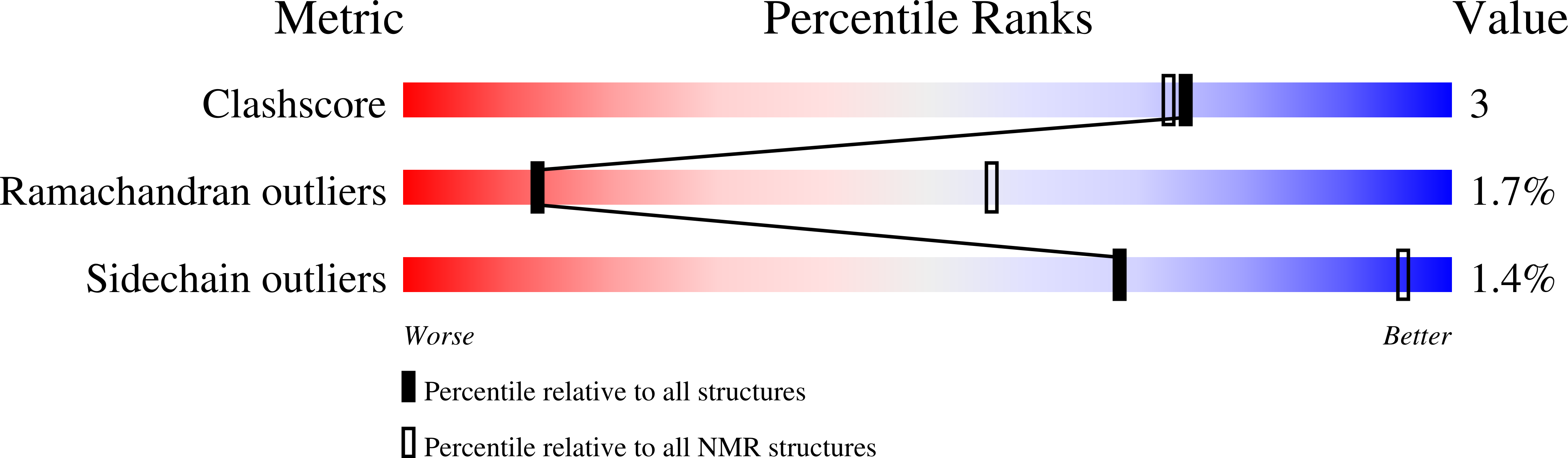Nucleation of a key beta-turn promotes cyclotide oxidative folding.
Tian, S., de Veer, S.J., Durek, T., Wang, C.K., Craik, D.J.(2024) J Biol Chem 300: 107125-107125
- PubMed: 38432638
- DOI: https://doi.org/10.1016/j.jbc.2024.107125
- Primary Citation of Related Structures:
8TYI - PubMed Abstract:
Cyclotides are plant-derived peptides characterized by a head-to-tail cyclic backbone and a cystine knot motif comprised of three disulfide bonds. Formation of this motif via in vitro oxidative folding can be challenging and can result in misfolded isomers with nonnative disulfide connectivities. Here, we investigated the effect of β-turn nucleation on cyclotide oxidative folding. Two types of β-turn mimics were grafted into kalata B1, individually replacing each of the four β-turns in the folded cyclotide. Insertion of d-Pro-Gly into loop 5 was beneficial to the folding of both cyclic kB1 and a linear form of the peptide. The linear grafted analog folded four-times faster in aqueous conditions than cyclic kB1 in optimized conditions. Additionally, the cyclic analogue folded without the need for redox agents by transitioning through a native-like intermediate that was on-pathway to product formation. Kalata B1 is from the Möbius subfamily of cyclotides. Grafting d-Pro-Gly into loop 5 of cyclotides from two other subfamilies also had a beneficial effect on folding. Our findings demonstrate the importance of a β-turn nucleation site for cyclotide oxidative folding, which could be adopted as a chemical strategy to improve the in vitro folding of diverse cystine-rich peptides.
Organizational Affiliation:
Institute for Molecular Bioscience, Australian Research Council Centre of Excellence for Innovations in Peptide and Protein Science, The University of Queensland, Brisbane, Queensland, Australia.