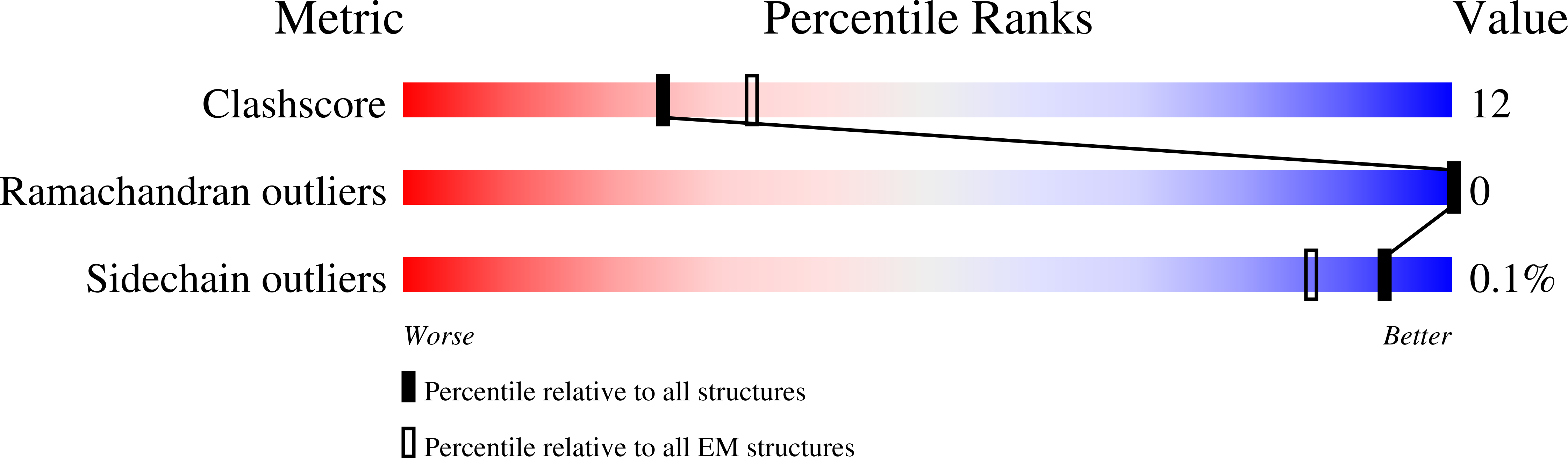Substrate recognition mechanism of the endoplasmic reticulum-associated ubiquitin ligase Doa10.
Wu, K., Itskanov, S., Lynch, D.L., Chen, Y., Turner, A., Gumbart, J.C., Park, E.(2024) Nat Commun 15: 2182-2182
- PubMed: 38467638
- DOI: https://doi.org/10.1038/s41467-024-46409-2
- Primary Citation of Related Structures:
8TQM - PubMed Abstract:
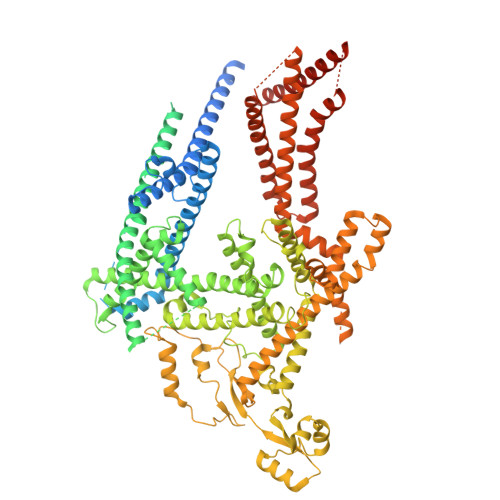
Doa10 (MARCHF6 in metazoans) is a large polytopic membrane-embedded E3 ubiquitin ligase in the endoplasmic reticulum (ER) that plays an important role in quality control of cytosolic and ER proteins. Although Doa10 is highly conserved across eukaryotes, it is not understood how Doa10 recognizes its substrates. Here, we define the substrate recognition mechanism of Doa10 by structural and functional analyses on Saccharomyces cerevisiae Doa10 and its model substrates. Cryo-EM analysis shows that Doa10 has unusual architecture with a large lipid-filled central cavity, and its conserved middle domain forms an additional water-filled lateral tunnel open to the cytosol. Our biochemical data and molecular dynamics simulations suggest that the entrance of the substrate's degron peptide into the lateral tunnel is required for efficient polyubiquitination. The N- and C-terminal membrane domains of Doa10 seem to form fence-like features to restrict polyubiquitination to those proteins that can access the central cavity and lateral tunnel. Our study reveals how extended hydrophobic sequences at the termini of substrate proteins are recognized by Doa10 as a signal for quality control.
Organizational Affiliation:
Department of Molecular and Cell Biology, University of California, Berkeley, CA, 94720, USA.