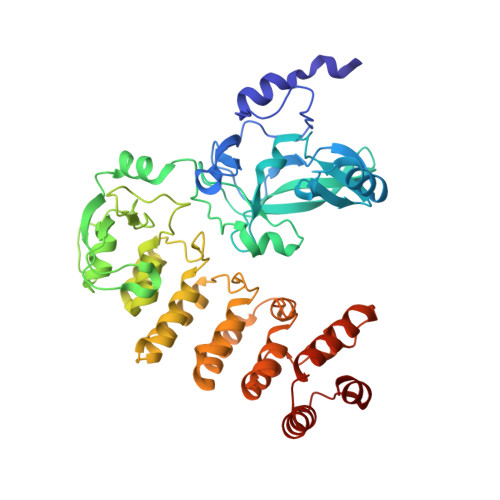
The SET and ankyrin domains of the secreted Legionella pneumophila histone methyltransferase work together to modify host chromatin.
Rolando, M., Wah Chung, I.Y., Xu, C., Gomez-Valero, L., England, P., Cygler, M., Buchrieser, C.(2023) mBio 14: e0165523-e0165523
- PubMed: 37795993
- DOI: https://doi.org/10.1128/mbio.01655-23
- Primary Citation of Related Structures:
8SWI - PubMed Abstract:
Legionella pneumophila is an intracellular bacterium responsible of Legionnaires' disease, a severe pneumonia that is often fatal when not treated promptly. The pathogen's ability to efficiently colonize the host resides in its ability to replicate intracellularly. Essential for intracellular replication is translocation of many different protein effectors via a specialized secretion system. One of them, called RomA, binds and directly modifies the host chromatin at a unique site (tri-methylation of lysine 14 of histone H3 [H3K14me]). However, the molecular mechanisms of binding are not known. Here, we resolve this question through structural characterization of RomA together with the H3 peptide. We specifically reveal an active role of the ankyrin repeats located in its C-terminal in the interaction with the histone H3 tail. Indeed, without the ankyrin domains, RomA loses its ability to act as histone methyltransferase. These results discover the molecular mechanisms by which a bacterial histone methyltransferase that is conserved in L. pneumophila strains acts to modify chromatin.
Organizational Affiliation:
Institut Pasteur, Université de Paris, Biologie des Bactéries Intracellulaires , Paris, France.