Crystal structure and interconversion of monomers and domain-swapped dimers of the walnut tree phytocystatin.
Simpson, G.A., Rezende, I.F., da Silva Peixoto, A., de Oliveira Soares, I.B., Barbosa, J.A.R.G., de Freitas, S.M., Valadares, N.F.(2023) Biochim Biophys Acta Proteins Proteom 1872: 140975-140975
- PubMed: 38056804
- DOI: https://doi.org/10.1016/j.bbapap.2023.140975
- Primary Citation of Related Structures:
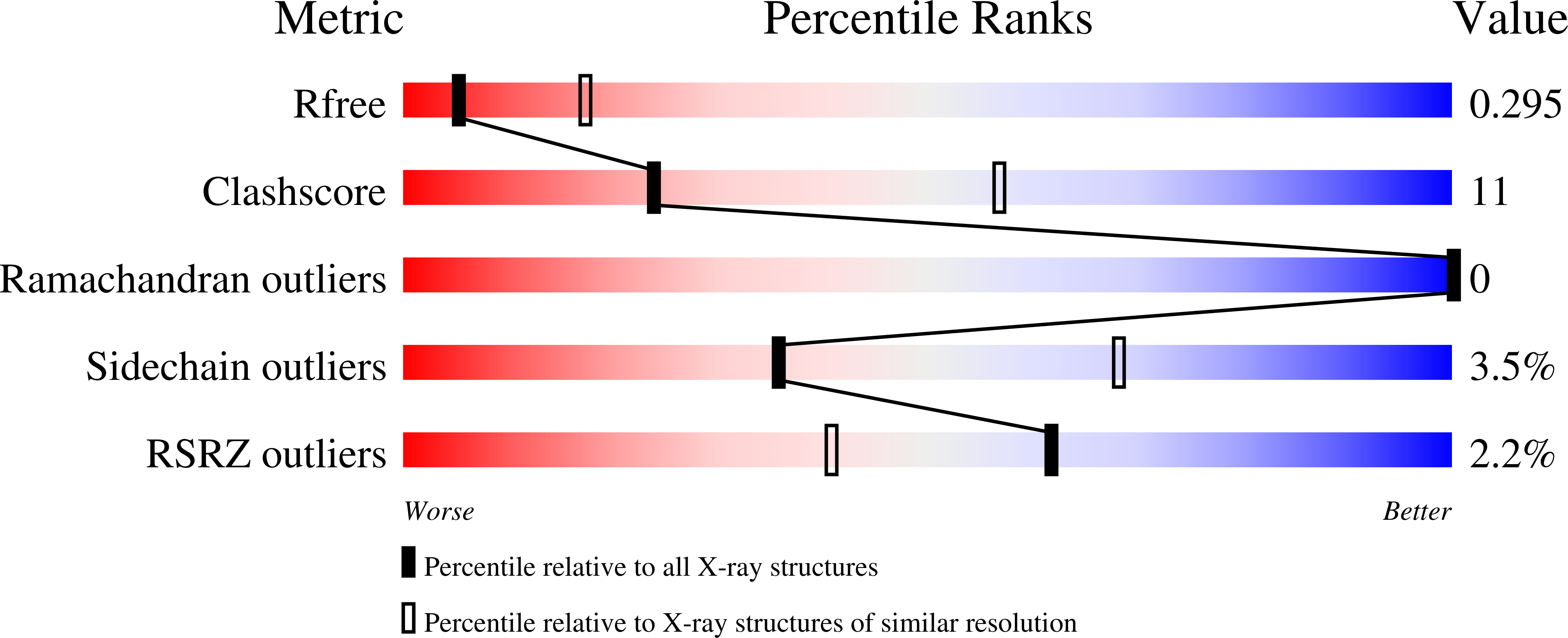
8SJ5 - PubMed Abstract:
Biotechnological applications of phytocystatins have garnered significant interest due to their potential applications in crop protection and improve crop resistance to abiotic stress factors. Cof1 and Wal1 are phytocystatins derived from Coffea arabica and Juglans regia, respectively. These plants hold significant economic value due to coffee's global demand and the walnut tree's production of valuable timber and widely consumed walnuts with culinary and nutritional benefits. The study involved the heterologous expression in E. coli Lemo 21(DE3), purification by immobilized metal ion affinity and size exclusion chromatography, and biophysical characterization of both phytocystatins, focusing on isolating and interconverting their monomers and dimers. The crystal structure of the domain-swapped dimer of Wal1 was determined revealing two domain-swapped dimers in the asymmetric unit, an arrangement reminiscent of the human cystatin C structure. Alphafold models of monomers and Alphafold-Multimer models of domain-swapped dimers of Cof1 and Wal1 were analyzed in the context of the crystal structure. The methodology and data presented here contribute to a deeper understanding of the oligomerization mechanisms of phytocystatins and their potential biotechnological applications in agriculture.
Organizational Affiliation:
Laboratory of Molecular Biophysics, Department of Cellular Biology, University of Brasília, Brasília, Brazil.