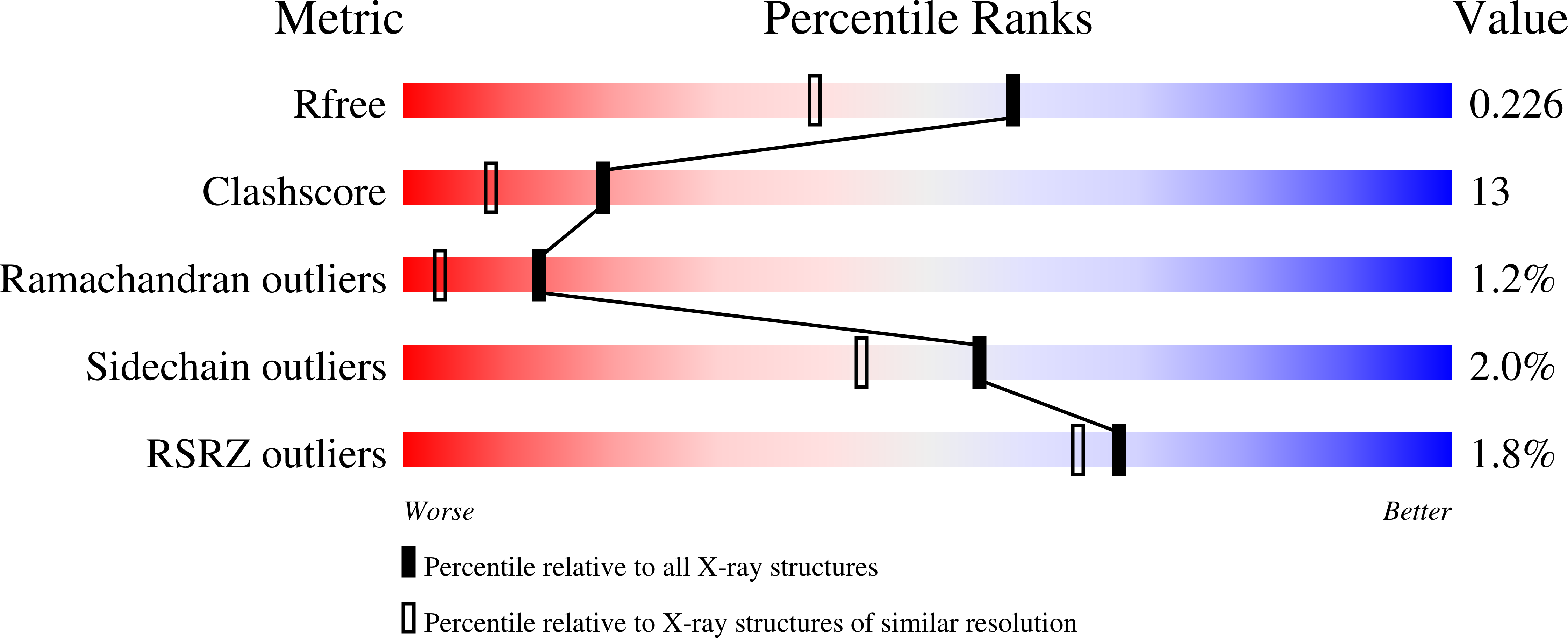
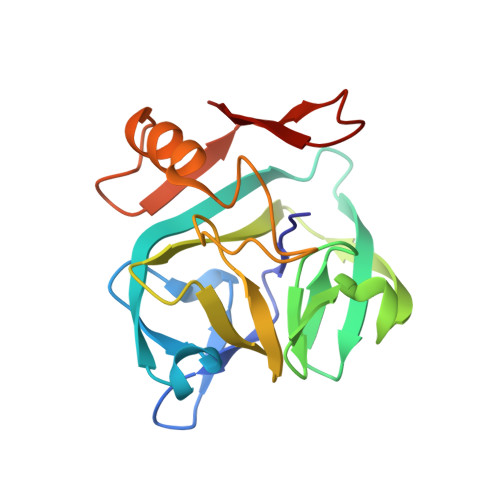
The three-dimensional structure of the Vint domain from Tetrahymena thermophila suggests a ligand-regulated cleavage mechanism by the HINT fold.
Iwai, H., Beyer, H.M., Johansson, J.E.M., Li, M., Wlodawer, A.(2024) FEBS Lett 598: 864-874
- PubMed: 38351630
- DOI: https://doi.org/10.1002/1873-3468.14817
- Primary Citation of Related Structures:
8R2C - PubMed Abstract:
Vint proteins have been identified in unicellular metazoans as a novel hedgehog-related gene family, merging the von Willebrand factor type A domain and the Hedgehog/INTein (HINT) domains. We present the first three-dimensional structure of the Vint domain from Tetrahymena thermophila corresponding to the auto-processing domain of hedgehog proteins, shedding light on the unique features, including an adduct recognition region (ARR). Our results suggest a potential binding between the ARR and sulfated glycosaminoglycans like heparin sulfate. Moreover, we uncover a possible regulatory role of the ARR in the auto-processing by Vint domains, expanding our understanding of the HINT domain evolution and their use in biotechnological applications. Vint domains might have played a crucial role in the transition from unicellular to multicellular organisms.
Organizational Affiliation:
Institute of Biotechnology, University of Helsinki, Finland.