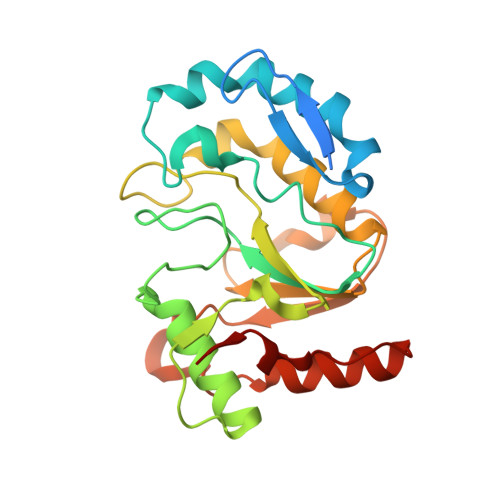
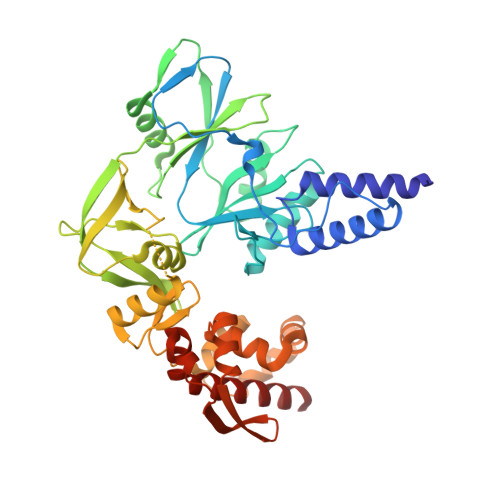
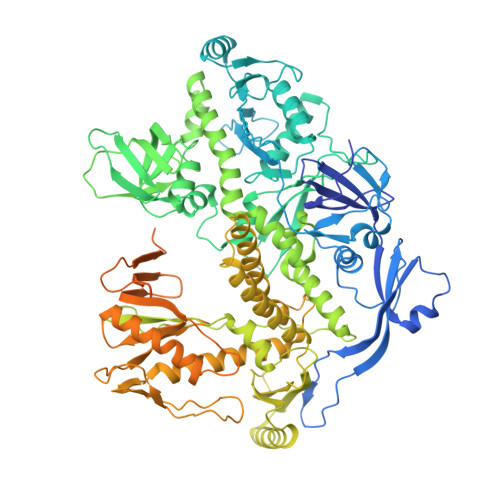
Structure and flexibility of the DNA polymerase holoenzyme of vaccinia virus.
Burmeister, W.P., Boutin, L., Balestra, A.C., Groger, H., Ballandras-Colas, A., Hutin, S., Kraft, C., Grimm, C., Bottcher, B., Fischer, U., Tarbouriech, N., Iseni, F.(2024) PLoS Pathog 20: e1011652-e1011652
- PubMed: 38768256
- DOI: https://doi.org/10.1371/journal.ppat.1011652
- Primary Citation of Related Structures:
8Q3R, 8QAM - PubMed Abstract:
The year 2022 was marked by the mpox outbreak caused by the human monkeypox virus (MPXV), which is approximately 98% identical to the vaccinia virus (VACV) at the sequence level with regard to the proteins involved in DNA replication. We present the production in the baculovirus-insect cell system of the VACV DNA polymerase holoenzyme, which consists of the E9 polymerase in combination with its co-factor, the A20-D4 heterodimer. This led to the 3.8 Å cryo-electron microscopy (cryo-EM) structure of the DNA-free form of the holoenzyme. The model of the holoenzyme was constructed from high-resolution structures of the components of the complex and the A20 structure predicted by AlphaFold 2. The structures do not change in the context of the holoenzyme compared to the previously determined crystal and NMR structures, but the E9 thumb domain became disordered. The E9-A20-D4 structure shows the same compact arrangement with D4 folded back on E9 as observed for the recently solved MPXV holoenzyme structures in the presence and the absence of bound DNA. A conserved interface between E9 and D4 is formed by a cluster of hydrophobic residues. Small-angle X-ray scattering data show that other, more open conformations of E9-A20-D4 without the E9-D4 contact exist in solution using the flexibility of two hinge regions in A20. Biolayer interferometry (BLI) showed that the E9-D4 interaction is indeed weak and transient in the absence of DNA although it is very important, as it has not been possible to obtain viable viruses carrying mutations of key residues within the E9-D4 interface.
Organizational Affiliation:
Institut de Biologie Structurale (IBS), Université Grenoble Alpes (UGA), Commissariat à l'Energie Atomique et aux Energies Alternatives (CEA), Centre National de la Recherche Scientifique (CNRS), Grenoble, France.