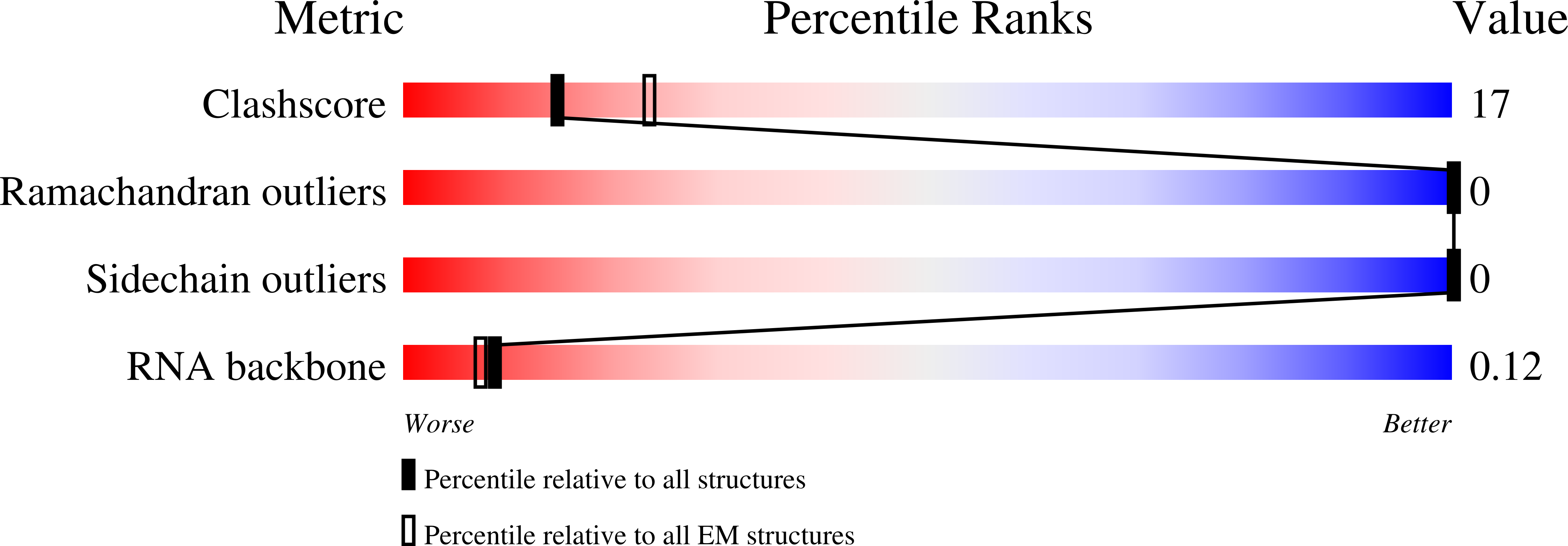
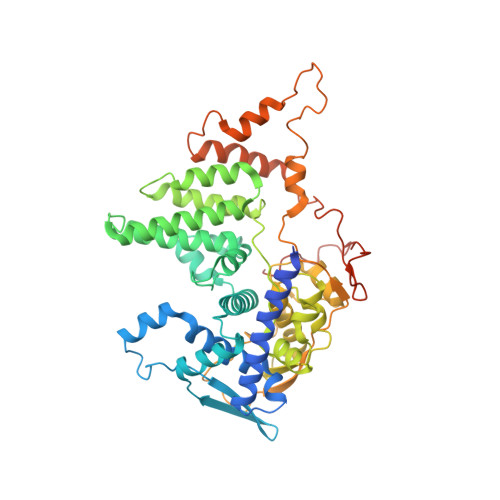
Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation.
Chenavier, F., Estrozi, L.F., Teulon, J.M., Zarkadas, E., Freslon, L.L., Pellequer, J.L., Ruigrok, R.W.H., Schoehn, G., Ballandras-Colas, A., Crepin, T.(2023) Sci Adv 9: eadj9974-eadj9974
- PubMed: 38100595
- DOI: https://doi.org/10.1126/sciadv.adj9974
- Primary Citation of Related Structures:
8PZP, 8PZQ - PubMed Abstract:
Influenza virus genome encapsidation is essential for the formation of a helical viral ribonucleoprotein (vRNP) complex composed of nucleoproteins (NP), the trimeric polymerase, and the viral genome. Although low-resolution vRNP structures are available, it remains unclear how the viral RNA is encapsidated and how NPs assemble into the helical filament specific of influenza vRNPs. In this study, we established a biological tool, the RNP-like particles assembled from recombinant influenza A virus NP and synthetic RNA, and we present the first subnanometric cryo-electron microscopy structure of the helical NP-RNA complex (8.7 to 5.3 Å). The helical RNP-like structure reveals a parallel double-stranded conformation, allowing the visualization of NP-NP and NP-RNA interactions. The RNA, located at the interface of neighboring NP protomers, interacts with conserved residues previously described as essential for the NP-RNA interaction. The NP undergoes conformational changes to enable RNA binding and helix formation. Together, our findings provide relevant insights for understanding the mechanism for influenza genome encapsidation.
Organizational Affiliation:
Univ. Grenoble Alpes, CNRS, CEA, IBS, F-38000, Grenoble, France.