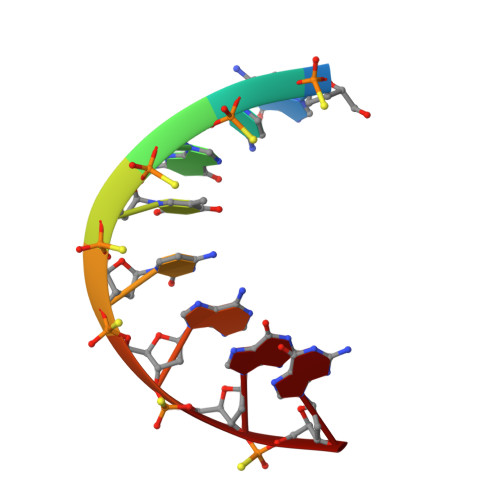
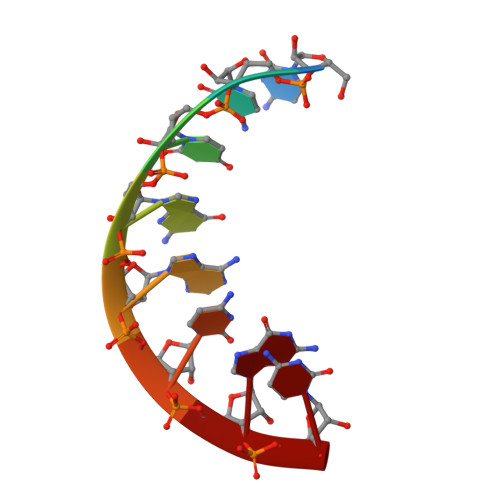
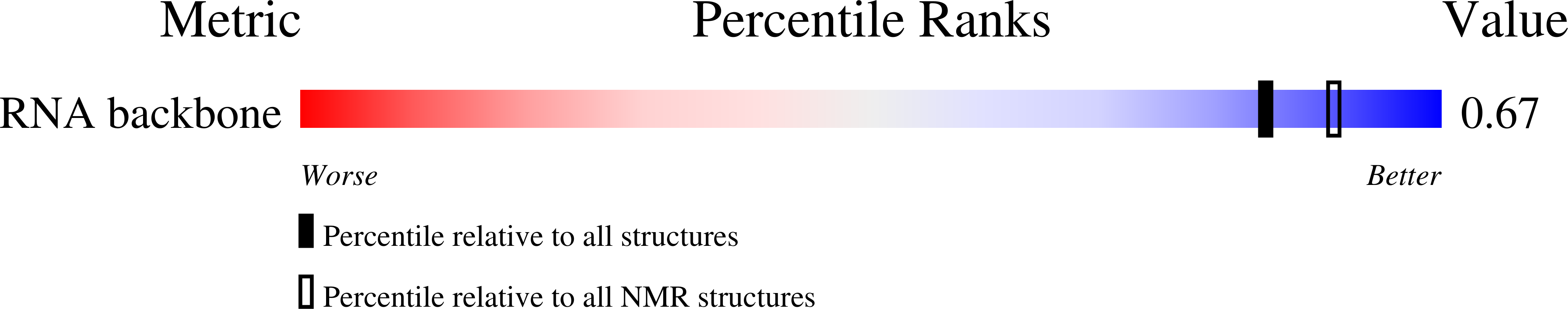Structure of a Stereoregular Phosphorothioate DNA/RNA Duplex
Bachelin, M., Hessler, G., Kurz, G., Hacia, J.G., Dervan, P.B., Kessler, H.(1998) Nat Struct Biol 5: 271-276
- PubMed: 9546216
- DOI: https://doi.org/10.1038/nsb0498-271
- Primary Citation of Related Structures:
8DRH, 8PSH - PubMed Abstract:
In this work, we present the first NMR solution structure of a DNA/RNA hybrid containing stereoregular Rp-phosphorothioate modifications of all DNA backbone linkages. The complex of the enzymatically synthesized phosphorothioate DNA octamer (all-Rp)-d(GCGTCAGG) and its complementary RNA r(CCUGACGC) was found to adopt an overall conformation within the A-form family. Most helical parameters and the sugar puckers of the DNA strand assume values intermediate between A- and B-form. The close structural similarity with the unmodified DNA/RNA hybrid of the same sequence may explain why both the natural and the sulfur-substituted complex can be recognized and digested by ribonuclease H.
Organizational Affiliation:
Institut für Organische Chemie und Biochemie, TU München, Garching, Germany.