Innate programmable DNA binding by CRISPR-Cas12m effectors enable efficient base editing.
Bigelyte, G., Duchovska, B., Zedaveinyte, R., Sasnauskas, G., Sinkunas, T., Dalgediene, I., Tamulaitiene, G., Silanskas, A., Kazlauskas, D., Valancauskas, L., Madariaga-Marcos, J., Seidel, R., Siksnys, V., Karvelis, T.(2024) Nucleic Acids Res 52: 3234-3248
- PubMed: 38261981
- DOI: https://doi.org/10.1093/nar/gkae016
- Primary Citation of Related Structures:
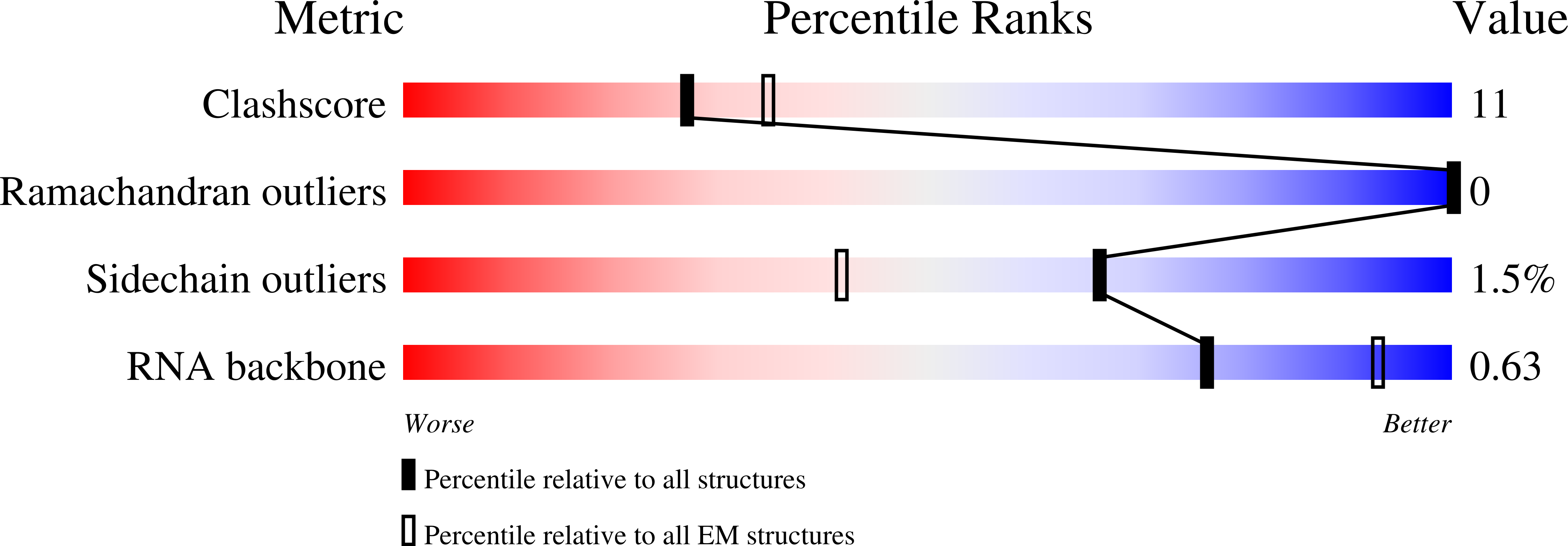
8PM4 - PubMed Abstract:
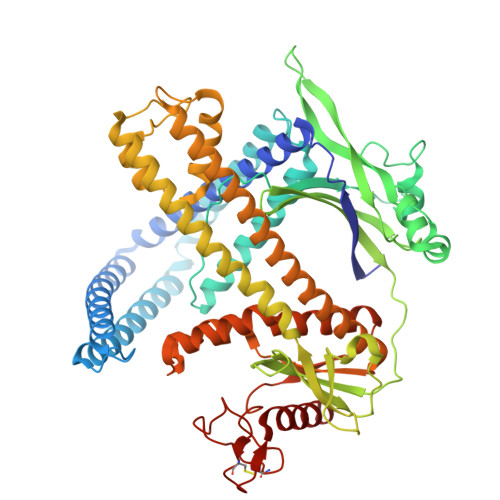
Cas9 and Cas12 nucleases of class 2 CRISPR-Cas systems provide immunity in prokaryotes through RNA-guided cleavage of foreign DNA. Here we characterize a set of compact CRISPR-Cas12m (subtype V-M) effector proteins and show that they provide protection against bacteriophages and plasmids through the targeted DNA binding rather than DNA cleavage. Biochemical assays suggest that Cas12m effectors can act as roadblocks inhibiting DNA transcription and/or replication, thereby triggering interference against invaders. Cryo-EM structure of Gordonia otitidis (Go) Cas12m ternary complex provided here reveals the structural mechanism of DNA binding ensuring interference. Harnessing GoCas12m innate ability to bind DNA target we fused it with adenine deaminase TadA-8e and showed an efficient A-to-G editing in Escherichia coli and human cells. Overall, this study expands our understanding of the functionally diverse Cas12 protein family, revealing DNA-binding dependent interference mechanism of Cas12m effectors that could be harnessed for engineering of compact base-editing tools.
Organizational Affiliation:
Institute of Biotechnology, Life Sciences Center, Vilnius University, Vilnius LT-10257, Lithuania.