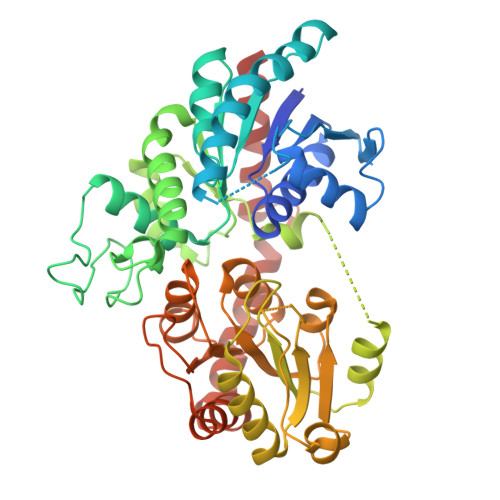
Molecular basis of ligand recognition specificity of flavone glucosyltransferases in Nemophila menziesii.
Murayama, K., Kato-Murayama, M., Hosaka, T., Okitsu, N., Tanaka, Y., Shirouzu, M.(2024) Arch Biochem Biophys 753: 109926-109926
- PubMed: 38346547
- DOI: https://doi.org/10.1016/j.abb.2024.109926
- Primary Citation of Related Structures:
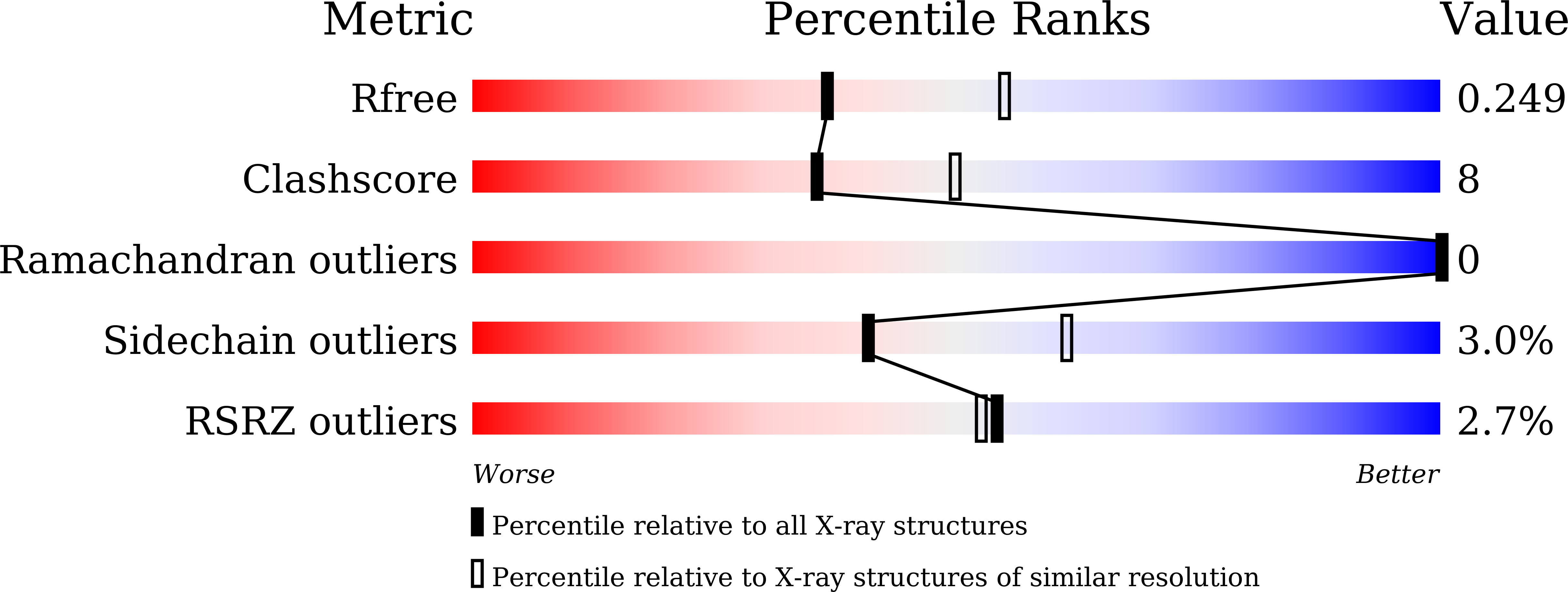
8I8Z, 8I90, 8I94 - PubMed Abstract:
Of the more than 100 families of glycosyltransferases, family 1 glycosyltransferases catalyze glycosylation using uridine diphosphate (UDP)-sugar as a sugar donor and are thus referred to as UDP-sugar:glycosyl transferases. The blue color of the Nemophila menziesii flower is derived from metalloanthocyanin, which consists of anthocyanin, flavone, and metal ions. Flavone 7-O-β-glucoside-4'-O-β-glucoside in the plant is sequentially biosynthesized from flavons by UDP-glucose:flavone 4'-O-glucosyltransferase (NmF4'GT) and UDP-glucose:flavone 4'-O-glucoside 7-O-glucosyltransferase (NmF4'G7GT). To identify the molecular mechanisms of glucosylation of flavone, the crystal structures of NmF4'G7GT in its apo form and in complex with UDP-glucose or luteolin were determined, and molecular structure prediction using AlphaFold2 was conducted for NmF4'GT. The crystal structures revealed that the size of the ligand-binding pocket and interaction environment for the glucose moiety at the pocket entrance plays a critical role in the substrate preference in NmF4'G7GT. The substrate specificity of NmF4'GT was examined by comparing its model structure with that of NmF4'G7GT. The structure of NmF4'GT may have a smaller acceptor pocket, leading to a substrate preference for non-glucosylated flavones (or flavone aglycones).
Organizational Affiliation:
Division of Biomedical Measurements and Diagnostics, Graduate School of Biomedical Engineering, Tohoku University, Sendai, 980-8575, Japan; Laboratory for Protein Functional and Structural Biology, RIKEN Center for Biosystems Dynamics Research, Yokohama, 230-0045, Japan.