A Tri-Enzyme Cascade for Efficient Production of L-2-Aminobutyrate from L-Threonine.
Li, X., Gao, C., Wei, W., Song, W., Meng, W., Liu, J., Chen, X., Gao, C., Guo, L., Liu, L., Wu, J.(2023) Chembiochem 24: e202300148-e202300148
- PubMed: 36946691
- DOI: https://doi.org/10.1002/cbic.202300148
- Primary Citation of Related Structures:
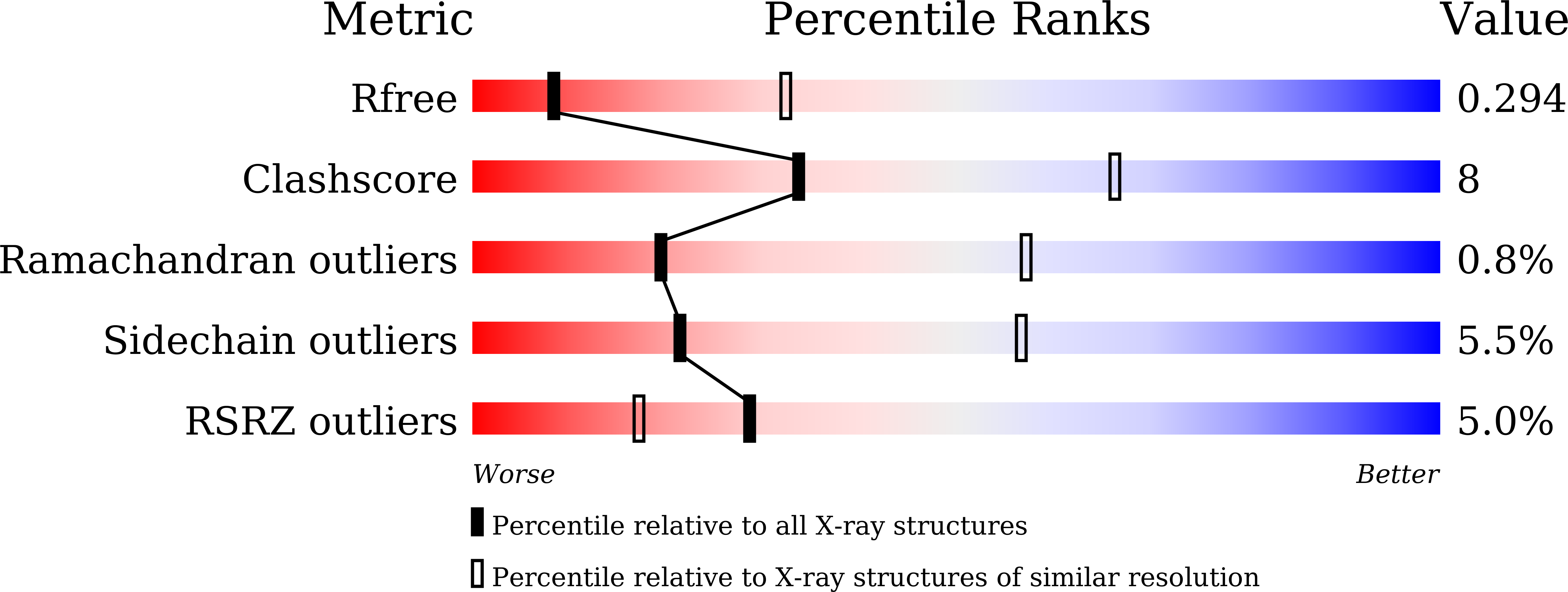
8HPE, 8HR6 - PubMed Abstract:
L-2-aminobutyrate (L-ABA) is an important chiral drug intermediate with a key role in modern medicinal chemistry. Here, we describe the development of an efficient method for the asymmetric synthesis of L-ABA in a tri-enzymatic cascade in Escherichia coli BL21 (DE3) using a cost-effective L-Thr. Low activity of leucine dehydrogenase from Bacillus thuringiensis (BtLDH) and unbalanced expression of enzymes in the cascade were major challenges. Mechanism-based protein engineering generated the optimal triple variant BtLDH M3 (A262S/V296C/P150M) with 20.7-fold increased specific activity and 9.6-fold increased k cat /K m compared with the wild type. Optimizing plasmids with different copy numbers regulated enzymatic expression, thereby increasing the activity ratio (0.3 : 1:0.6) of these enzymes in vivo close to the optimal ratio (0.4 : 1 : 1) in vitro. Importing the optimal triple mutant BtLDH M3 into our constructed pathway in vivo and optimization of transformation conditions achieved one-pot conversion of L-Thr to 130.2 g/L L-ABA, with 95 % conversion, 99 % e.e. and 10.9 g L -1 h -1 productivity (the highest to date) in 12 h on a 500 mL scale. These results describe a potential biosynthesis approach for the industrial production of L-ABA.
Organizational Affiliation:
School of Life Sciences and Health Engineering, Jiangnan University, Wuxi, 214122, China.