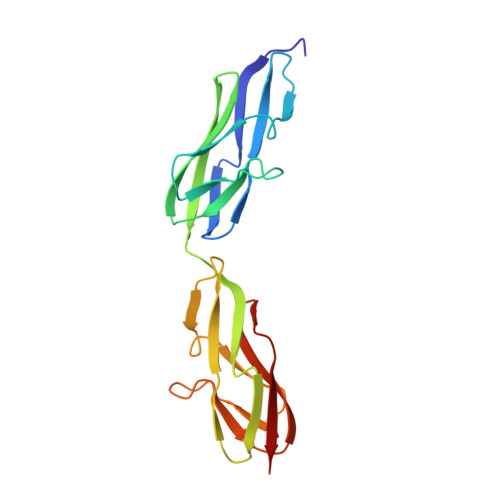
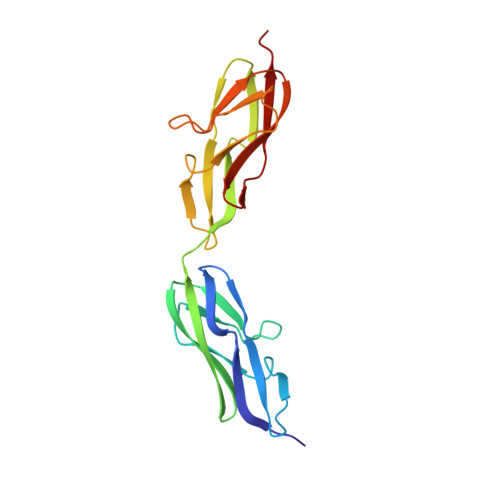Crystal structure of a variable region segment of Leptospira host-interacting outer surface protein, LigA, reveals the orientation of Ig-like domains.
Kumar, P., Vyas, P., Faisal, S.M., Chang, Y.F., Akif, M.(2023) Int J Biol Macromol 244: 125445-125445
- PubMed: 37336372
- DOI: https://doi.org/10.1016/j.ijbiomac.2023.125445
- Primary Citation of Related Structures:
8GYR - PubMed Abstract:
Leptospiral immunoglobulin-like (Lig) protein family is a surface-exposed protein from the pathogenic Leptospira. The Lig protein family has been identified as an essential virulence factor of L. interrogan. One of the family members, LigA, contains 13 homologous tandem repeats of bacterial Ig-like (Big) domains in its extracellular portion. It is crucial in binding with the host's Extracellular matrices (ECM) and complement factors. However, its vital role in the invasion and evasion of pathogenic Leptospira, structural details, and domain organization of the extracellular portion of this protein are not explored thoroughly. Here, we described the first high-resolution crystal structure of a variable region segment (LigA8-9) of LigA at 1.87 Å resolution. The structure showed some remarkably distinctive aspects compared with other closely related Immunoglobulin domains. The structure illustrated the relative orientation of two domains and highlighted the role of the linker region in the domain orientation. We also observed an apparent electron density of Ca 2+ ions coordinated with a proper interacting geometry within the protein. Molecular dynamic simulations demonstrated the involvement of a linker salt bridge in providing rigidity between the two domains. Our study proposes an overall arrangement of Ig-like domains in the LigA protein. The structural understanding of the extracellular portion of LigA and its interaction with the ECM provides insight into developing new therapeutics directed toward leptospirosis.
Organizational Affiliation:
Laboratory of Structural Biology, Department of Biochemistry, School of Life Sciences, University of Hyderabad, Gachibowli, Hyderabad, India.