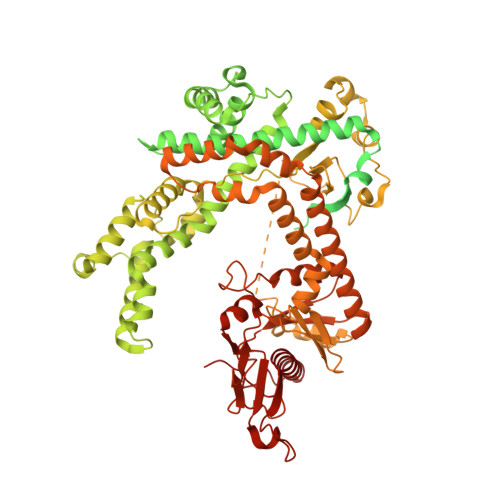
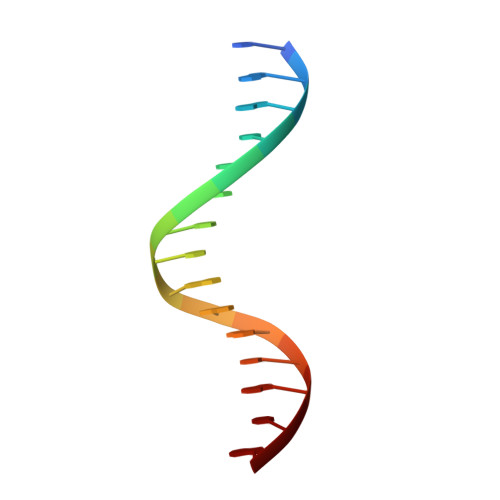
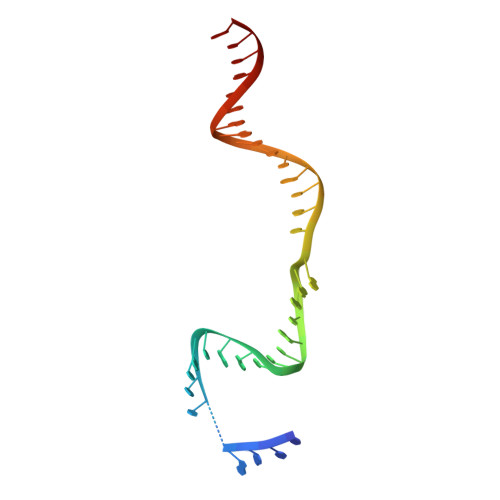
Fanzor is a eukaryotic programmable RNA-guided endonuclease.
Saito, M., Xu, P., Faure, G., Maguire, S., Kannan, S., Altae-Tran, H., Vo, S., Desimone, A., Macrae, R.K., Zhang, F.(2023) Nature 620: 660-668
- PubMed: 37380027
- DOI: https://doi.org/10.1038/s41586-023-06356-2
- Primary Citation of Related Structures:
8GKH - PubMed Abstract:
RNA-guided systems, which use complementarity between a guide RNA and target nucleic acid sequences for recognition of genetic elements, have a central role in biological processes in both prokaryotes and eukaryotes. For example, the prokaryotic CRISPR-Cas systems provide adaptive immunity for bacteria and archaea against foreign genetic elements. Cas effectors such as Cas9 and Cas12 perform guide-RNA-dependent DNA cleavage 1 . Although a few eukaryotic RNA-guided systems have been studied, including RNA interference 2 and ribosomal RNA modification 3 , it remains unclear whether eukaryotes have RNA-guided endonucleases. Recently, a new class of prokaryotic RNA-guided systems (termed OMEGA) was reported 4,5 . The OMEGA effector TnpB is the putative ancestor of Cas12 and has RNA-guided endonuclease activity 4,6 . TnpB may also be the ancestor of the eukaryotic transposon-encoded Fanzor (Fz) proteins 4,7 , raising the possibility that eukaryotes are also equipped with CRISPR-Cas or OMEGA-like programmable RNA-guided endonucleases. Here we report the biochemical characterization of Fz, showing that it is an RNA-guided DNA endonuclease. We also show that Fz can be reprogrammed for human genome engineering applications. Finally, we resolve the structure of Spizellomyces punctatus Fz at 2.7 Å using cryogenic electron microscopy, showing the conservation of core regions among Fz, TnpB and Cas12, despite diverse cognate RNA structures. Our results show that Fz is a eukaryotic OMEGA system, demonstrating that RNA-guided endonucleases are present in all three domains of life.
Organizational Affiliation:
Broad Institute of MIT and Harvard, Cambridge, MA, USA.