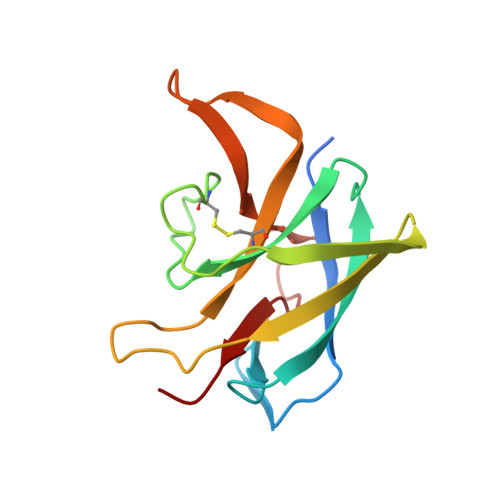
Dimerization of the CNNM extracellular domain.
Shahsavan, A., Lee, E.L., Illes, K., Kozlov, G., Gehring, K.(2024) Protein Sci 33: e4860-e4860
- PubMed: 38149326
- DOI: https://doi.org/10.1002/pro.4860
- Primary Citation of Related Structures:
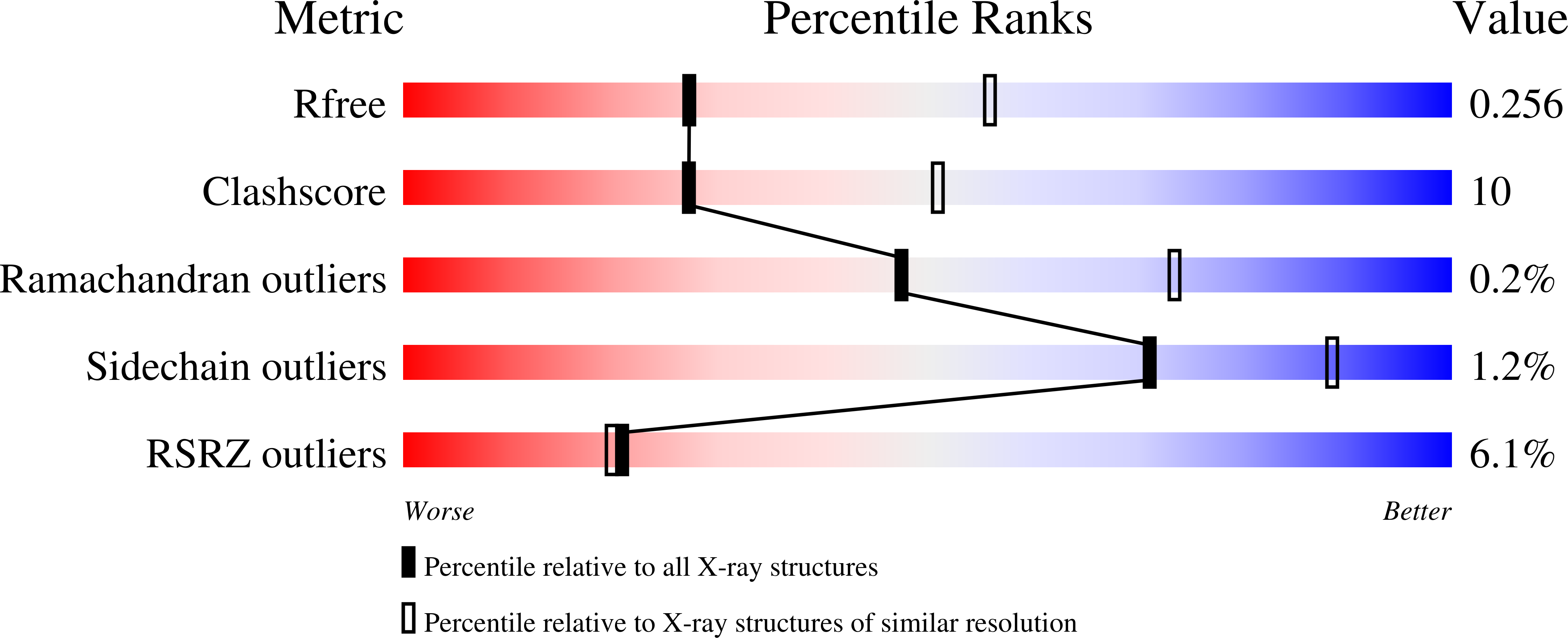
8GK6 - PubMed Abstract:
Cystathionine-
$$ \beta $$ -synthase (CBS)-pair domain divalent metal cation transport mediators (CNNMs) are an evolutionarily conserved family of magnesium transporters. They mediate magnesium homeostasis directly by transport of Mg 2+ ions and indirectly by regulation of the transient receptor potential ion channel subfamily M member 7 (TRPM7). Here, we report the crystal structure of the extracellular domain of tapeworm CNNM4. The domain forms a dimer of immunoglobulin-like (Ig-like) folds with electron density observed for three glycosylation sites. Analytical ultracentrifugation confirms that mutations in the extracellular domain of human CNNM4 prevent its dimerization. An analogous mutation in mouse CNNM2 impairs its activity in a cellular assay of Mg 2+ transport.
Organizational Affiliation:
Department of Biochemistry & Centre de recherche en biologie structurale, McGill University, Montreal, Canada.