Structural basis for flagellin-induced NAIP5 activation.
Paidimuddala, B., Cao, J., Zhang, L.(2023) Sci Adv 9: eadi8539-eadi8539
- PubMed: 38055825
- DOI: https://doi.org/10.1126/sciadv.adi8539
- Primary Citation of Related Structures:
8FML - PubMed Abstract:
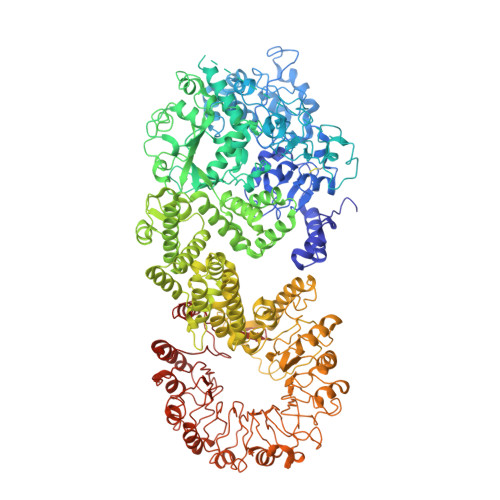
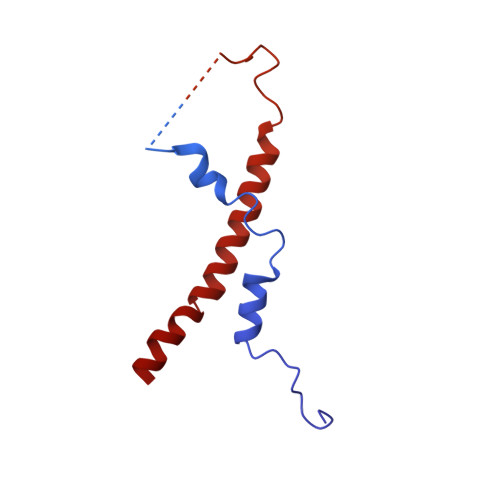
The NAIP (NLR family apoptosis inhibitory protein)/NLRC4 (NLR family CARD containing protein 4) inflammasome senses Gram-negative bacterial ligand. In the ligand-bound state, the winged helix domain of NAIP forms a steric clash with NLRC4 to open it up. However, how ligand binding activates NAIP is less clear. Here, we investigated the dynamics of the ligand-binding region of inactive NAIP5 and solved the cryo-EM structure of NAIP5 in complex with its specific ligand, FliC from flagellin, at 2.9-Å resolution. The structure revealed a "trap and lock" mechanism in FliC recognition, whereby FliC-D0 C is first trapped by the hydrophobic pocket of NAIP5, then locked in the binding site by ID (insertion domain) and C-terminal tail of NAIP5. The FliC-D0 N domain further inserts into ID to stabilize the complex. According to this mechanism, FliC triggers the conformational change of NAIP5 by bringing multiple flexible domains together.
Organizational Affiliation:
Department of Chemical Physiology and Biochemistry, Oregon Health and Science University, Portland, OR 97239, USA.