Crystal Structure of Enterovirus 68 3C Protease inactive mutant C147A at 1.8 Angstroms.
Azzolino, V.N., Shaqra, A.M., Schiffer, C.A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 3C Protease | 183 | enterovirus D68 | Mutation(s): 1 | 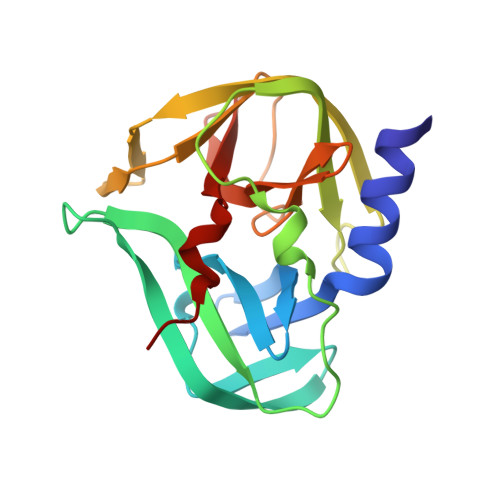 | |
UniProt | |||||
Find proteins for A0A097BW12 (Human enterovirus D68) Explore A0A097BW12 Go to UniProtKB: A0A097BW12 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A097BW12 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 39.166 | α = 90 |
| b = 102.109 | β = 109.88 |
| c = 41.946 | γ = 90 |
| Software Name | Purpose |
|---|---|
| phenix.refine | refinement |
| PHENIX | refinement |
| Coot | model building |
| CrysalisPro | data collection |
| CrysalisPro | data reduction |
| CrysalisPro | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other private | United States | -- |