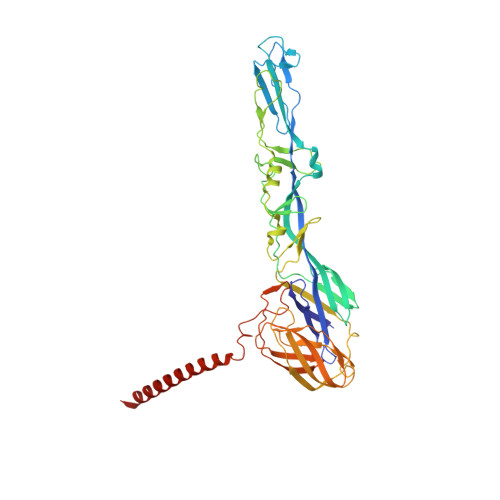
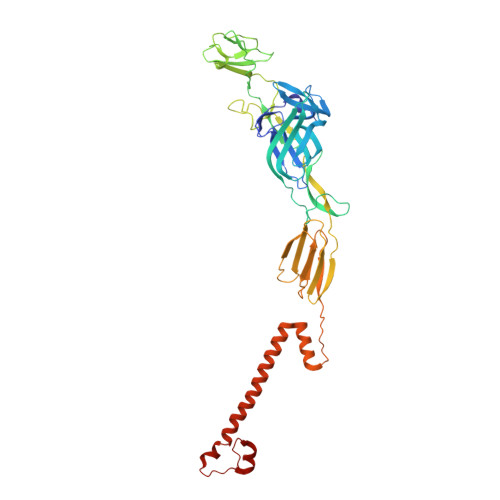
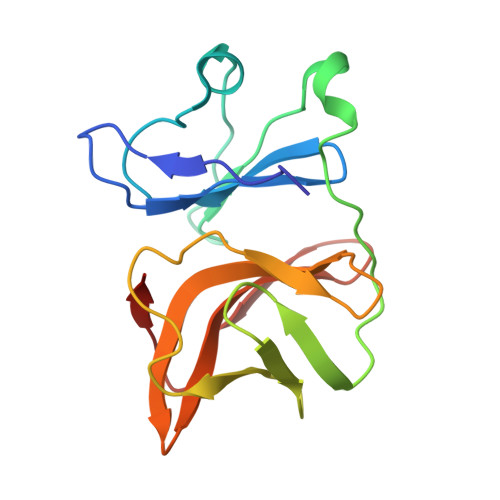
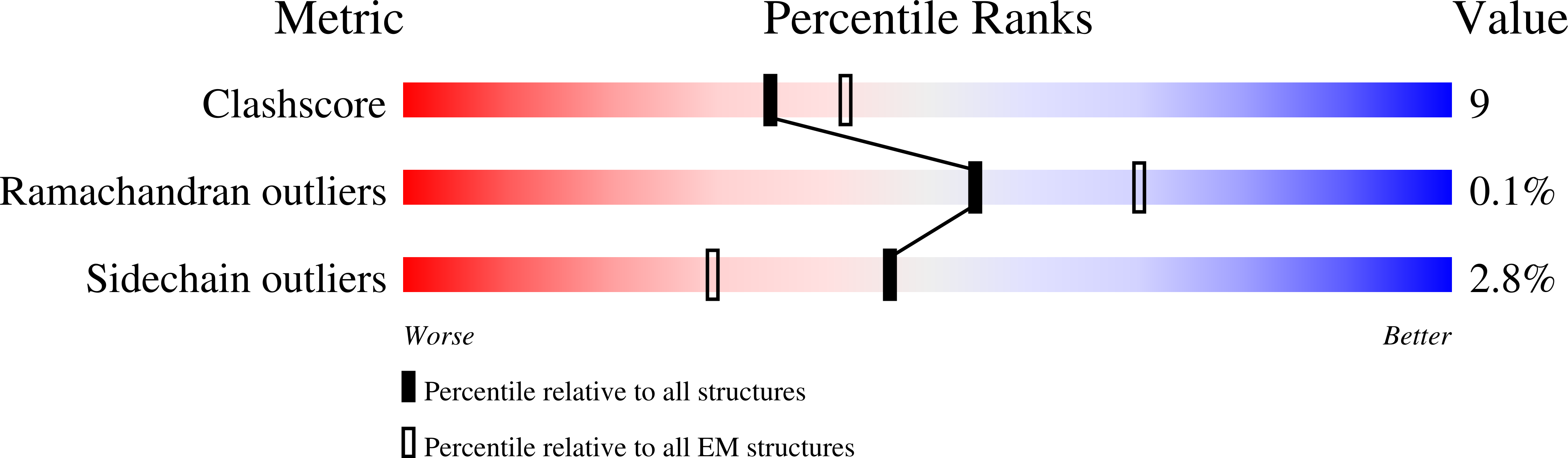Cryogenic electron microscopy and tomography reveal imperfect icosahedral symmetry in alphaviruses.
Chmielewski, D., Su, G.C., Kaelber, J.T., Pintilie, G.D., Chen, M., Jin, J., Auguste, A.J., Chiu, W.(2024) PNAS Nexus 3: pgae102-pgae102
- PubMed: 38525304
- DOI: https://doi.org/10.1093/pnasnexus/pgae102
- Primary Citation of Related Structures:
8FCG - PubMed Abstract:
Alphaviruses are spherical, enveloped RNA viruses with two-layered icosahedral architecture. The structures of many alphaviruses have been studied using cryogenic electron microscopy (cryo-EM) reconstructions, which impose icosahedral symmetry on the viral particles. Using cryogenic electron tomography (cryo-ET), we revealed a polarized symmetry defect in the icosahedral lattice of Chikungunya virus (CHIKV) in situ, similar to the late budding particles, suggesting the inherent imperfect symmetry originates from the final pinch-off of assembled virions. We further demonstrated this imperfect symmetry is also present in in vitro purified CHIKV and Mayaro virus, another arthritogenic alphavirus. We employed a subparticle-based single-particle analysis protocol to circumvent the icosahedral imperfection and boosted the resolution of the structure of the CHIKV to ∼3 Å resolution, which revealed detailed molecular interactions between glycoprotein E1-E2 heterodimers in the transmembrane region and multiple lipid-like pocket factors located in a highly conserved hydrophobic pocket. This complementary use of in situ cryo-ET and single-particle cryo-EM approaches provides a more precise structural description of near-icosahedral viruses and valuable insights to guide the development of structure-based antiviral therapies against alphaviruses.
Organizational Affiliation:
Biophysics Graduate Program, Stanford University, Stanford, CA 94305, USA.