Structural basis of Janus kinase trans-activation.
Caveney, N.A., Saxton, R.A., Waghray, D., Glassman, C.R., Tsutsumi, N., Hubbard, S.R., Garcia, K.C.(2023) Cell Rep 42: 112201-112201
- PubMed: 36867534
- DOI: https://doi.org/10.1016/j.celrep.2023.112201
- Primary Citation of Related Structures:
8EWY - PubMed Abstract:
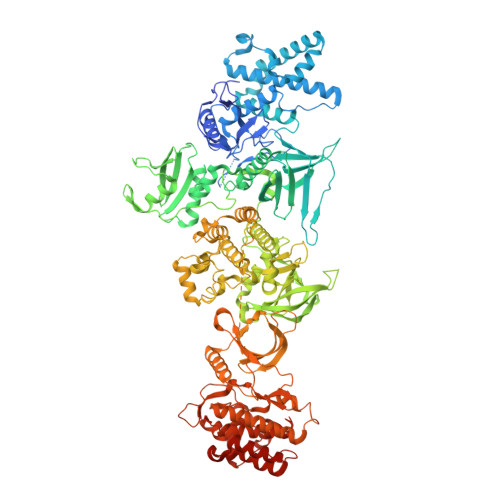
Janus kinases (JAKs) mediate signal transduction downstream of cytokine receptors. Cytokine-dependent dimerization is conveyed across the cell membrane to drive JAK dimerization, trans-phosphorylation, and activation. Activated JAKs in turn phosphorylate receptor intracellular domains (ICDs), resulting in the recruitment, phosphorylation, and activation of signal transducer and activator of transcription (STAT)-family transcription factors. The structural arrangement of a JAK1 dimer complex with IFNλR1 ICD was recently elucidated while bound by stabilizing nanobodies. While this revealed insights into the dimerization-dependent activation of JAKs and the role of oncogenic mutations in this process, the tyrosine kinase (TK) domains were separated by a distance not compatible with the trans-phosphorylation events between the TK domains. Here, we report the cryoelectron microscopy structure of a mouse JAK1 complex in a putative trans-activation state and expand these insights to other physiologically relevant JAK complexes, providing mechanistic insight into the crucial trans-activation step of JAK signaling and allosteric mechanisms of JAK inhibition.
Organizational Affiliation:
Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA 94305, USA.