Preclinical characterization and clinical translation of pharmacodynamic markers for MK-5890: a human CD27 activating antibody for cancer immunotherapy.
Guelen, L., Fischmann, T.O., Wong, J., Mauze, S., Guadagnoli, M., Babala, N., Wagenaars, J., Juan, V., Rosen, D., Prosise, W., Habraken, M., Lodewijks, I., Gu, D., Stammen-Vogelzangs, J., Yu, Y., Baker, J., Lutje Hulsik, D., Driessen-Engels, L., Malashock, D., Kreijtz, J., Bertens, A., de Vries, E., Bovens, A., Bramer, A., Zhang, Y., Wnek, R., Troth, S., Chartash, E., Dobrenkov, K., Sadekova, S., van Elsas, A., Cheung, J.K., Fayadat-Dilman, L., Borst, J., Beebe, A.M., Van Eenennaam, H.(2022) J Immunother Cancer 10
- PubMed: 36100308
- DOI: https://doi.org/10.1136/jitc-2022-005049
- Primary Citation of Related Structures:
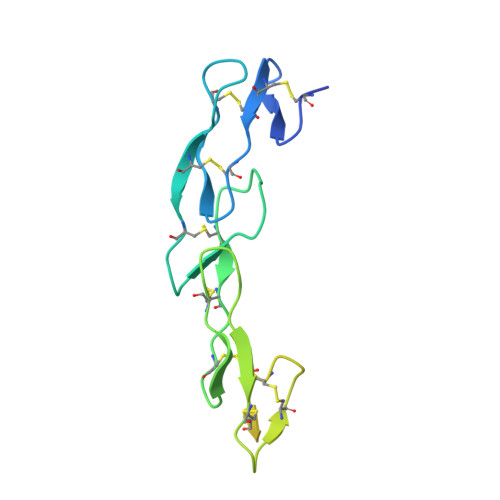
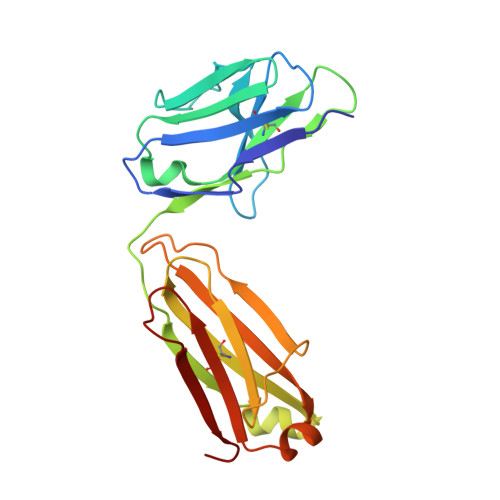
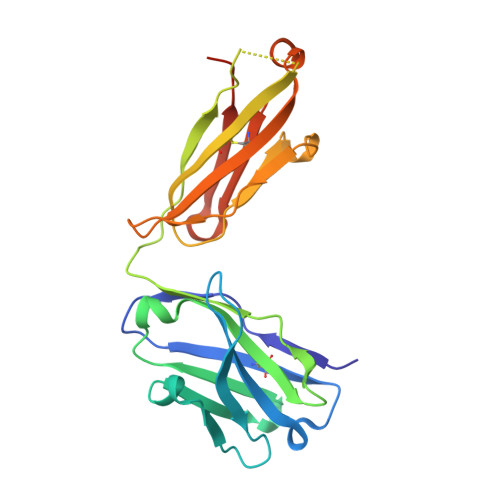
8DS5 - PubMed Abstract:
Immune checkpoint inhibitors (ICI) have radically changed cancer therapy, but most patients with cancer are unresponsive or relapse after treatment. MK-5890 is a CD27 agonist antibody intended to complement ICI therapy. CD27 is a member of the tumor necrosis factor receptor superfamily that plays a critical role in promoting responses of T cells, B cells and NK cells. Anti-CD27 antibodies were generated and selected for agonist activity using NF-кB luciferase reporter assays. Antibodies were humanized and characterized for agonism using in vitro T-cell proliferation assays. The epitope recognized on CD27 by MK-5890 was established by X-ray crystallography. Anti-tumor activity was evaluated in a human CD27 knock-in mouse. Preclinical safety was tested in rhesus monkeys. Pharmacodynamic properties were examined in mouse, rhesus monkeys and a phase 1 dose escalation clinical study in patients with cancer. Humanized anti-CD27 antibody MK-5890 (hIgG1) was shown to bind human CD27 on the cell surface with sub-nanomolar potency and to partially block binding to its ligand, CD70. Crystallization studies revealed that MK-5890 binds to a unique epitope in the cysteine-rich domain 1 (CRD1). MK-5890 activated CD27 expressed on 293T NF-κB luciferase reporter cells and, conditional on CD3 stimulation, in purified CD8+ T cells without the requirement of crosslinking. Functional Fc-receptor interaction was required to activate CD8+ T cells in an ex vivo tumor explant system and to induce antitumor efficacy in syngeneic murine subcutaneous tumor models. MK-5890 had monotherapy efficacy in these models and enhanced efficacy of PD-1 blockade. MK-5890 reduced in an isotype-dependent and dose-dependent manner circulating, but not tumor-infiltrating T-cell numbers in these mouse models. In rhesus monkey and human patients, reduction in circulating T cells was transient and less pronounced than in mouse. MK-5890 induced transient elevation of chemokines MCP-1, MIP-1α, and MIP-1β in the serum of mice, rhesus monkeys and patients with cancer. MK-5890 was well tolerated in rhesus monkeys and systemic exposure to MK-5890 was associated with CD27 occupancy at all doses. MK-5890 is a novel CD27 agonistic antibody with the potential to complement the activity of PD-1 checkpoint inhibition in cancer immunotherapy and is currently undergoing clinical evaluation.
Organizational Affiliation:
BioNovion/Aduro Biotech Europe, Oss, The Netherlands.