Bioinformatics of cyanophycin metabolism genes and characterization of promiscuous isoaspartyl dipeptidases that catalyze the final step of cyanophycin degradation.
Sharon, I., Schmeing, T.M.(2023) Sci Rep 13: 8314-8314
- PubMed: 37221236
- DOI: https://doi.org/10.1038/s41598-023-34587-w
- Primary Citation of Related Structures:
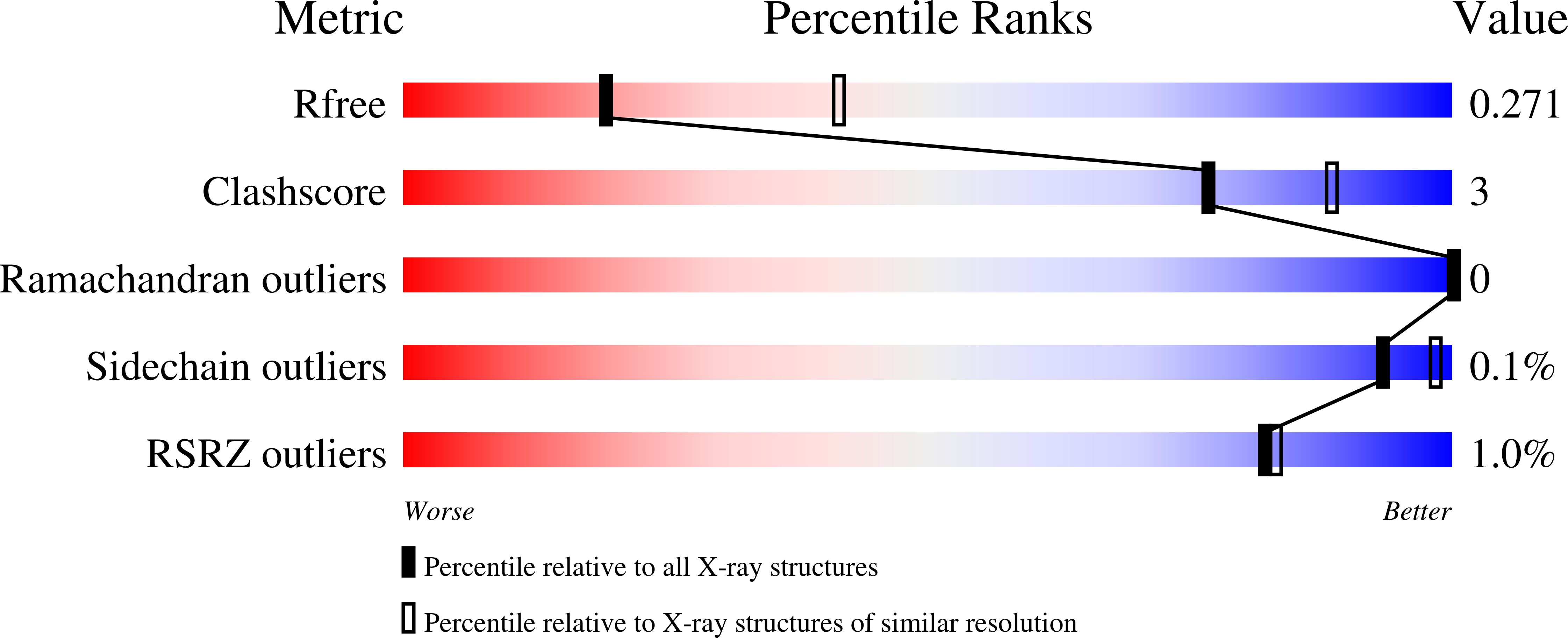
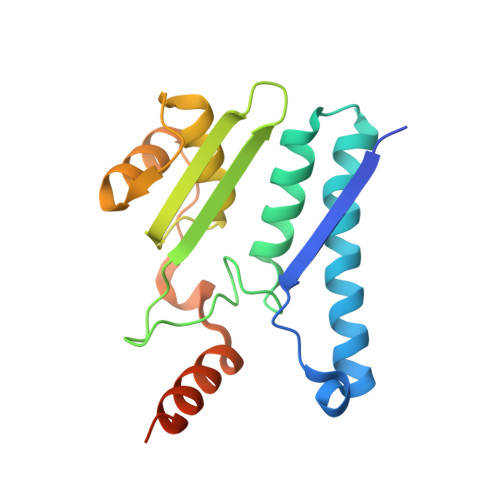
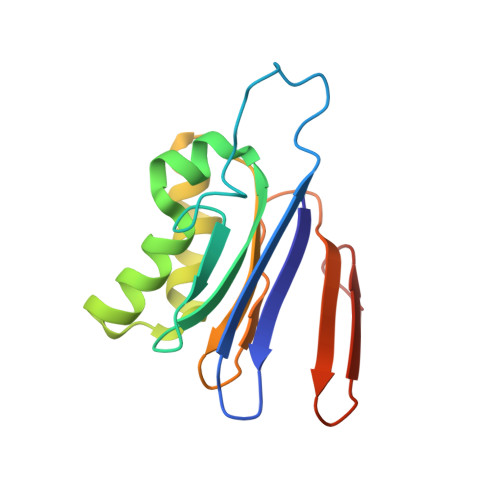
8DQM, 8DQN - PubMed Abstract:
Cyanophycin is a bacterial biopolymer used for storage of fixed nitrogen. It is composed of a backbone of L-aspartate residues with L-arginines attached to each of their side chains. Cyanophycin is produced by cyanophycin synthetase 1 (CphA1) using Arg, Asp and ATP, and is degraded in two steps. First, cyanophycinase breaks down the backbone peptide bonds, releasing β-Asp-Arg dipeptides. Then, these dipeptides are broken down into free Asp and Arg by enzymes with isoaspartyl dipeptidase activity. Two bacterial enzymes are known to possess promiscuous isoaspartyl dipeptidase activity: isoaspartyl dipeptidase (IadA) and isoaspartyl aminopeptidase (IaaA). We performed a bioinformatic analysis to investigate whether genes for cyanophycin metabolism enzymes cluster together or are spread around the microbial genomes. Many genomes showed incomplete contingents of known cyanophycin metabolizing genes, with different patterns in various bacterial clades. Cyanophycin synthetase and cyanophycinase are usually clustered together when recognizable genes for each are found within a genome. Cyanophycinase and isoaspartyl dipeptidase genes typically cluster within genomes lacking cphA1. About one-third of genomes with genes for CphA1, cyanophycinase and IaaA show these genes clustered together, while the proportion is around one-sixth for CphA1, cyanophycinase and IadA. We used X-ray crystallography and biochemical studies to characterize an IadA and an IaaA from two such clusters, in Leucothrix mucor and Roseivivax halodurans, respectively. The enzymes retained their promiscuous nature, showing that being associated with cyanophycin-related genes did not make them specific for β-Asp-Arg dipeptides derived from cyanophycin degradation.
Organizational Affiliation:
Department of Biochemistry and Centre de recherche en biologie structurale, McGill University, Montréal, QC, H3G 0B1, Canada.