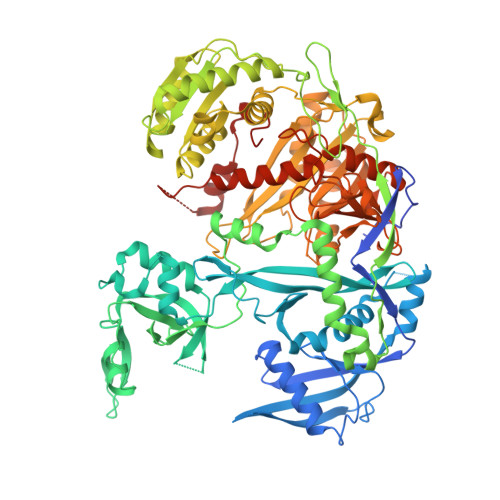
A tiny loop in the Argonaute PIWI domain tunes small RNA seed strength.
Xiao, Y., Liu, T.M., MacRae, I.J.(2023) EMBO Rep 24: e55806-e55806
- PubMed: 37082939
- DOI: https://doi.org/10.15252/embr.202255806
- Primary Citation of Related Structures:
8D6J, 8D71 - PubMed Abstract:
Argonaute (AGO) proteins use microRNAs (miRNAs) and small interfering RNAs (siRNAs) as guides to regulate gene expression in plants and animals. AGOs that use miRNAs in bilaterian animals recognize short (6-8 nt.) elements complementary to the miRNA seed region, enabling each miRNA to interact with hundreds of otherwise unrelated targets. By contrast, AGOs that use miRNAs in plants employ longer (> 13 nt.) recognition elements such that each miRNA silences a small number of physiologically related targets. Here, we show that this major functional distinction depends on a minor structural difference between plant and animal AGO proteins: a 9-amino acid loop in the PIWI domain. Swapping the PIWI loop from human Argonaute2 (HsAGO2) into Arabidopsis Argonaute10 (AtAGO10) increases seed strength, resulting in animal-like miRNA targeting. Conversely, swapping the plant PIWI loop into HsAGO2 reduces seed strength and accelerates the turnover of cleaved targets. The loop-swapped HsAGO2 silences targets more potently, with reduced miRNA-like targeting, than wild-type HsAGO2 in mammalian cells. Thus, tiny structural differences can tune the targeting properties of AGO proteins for distinct biological roles.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA, USA.