Conformational coupling of the sialic acid TRAP transporter HiSiaQM with its substrate binding protein HiSiaP.
Peter, M.F., Ruland, J.A., Kim, Y., Hendricks, P., Schneberger, N., Siebrasse, J.P., Thomas, G.H., Kubitscheck, U., Hagelueken, G.(2024) Nat Commun 15: 217-217
- PubMed: 38191530
- DOI: https://doi.org/10.1038/s41467-023-44327-3
- Primary Citation of Related Structures:
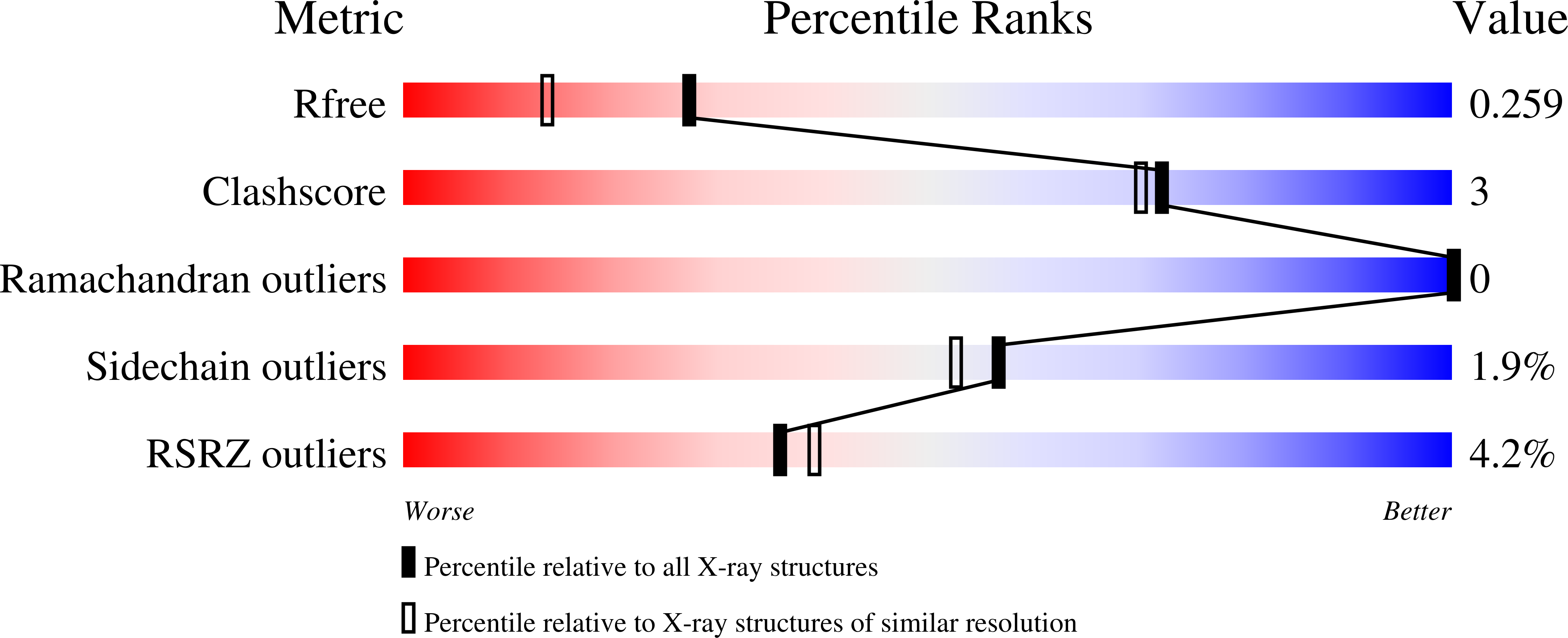
8CP7 - PubMed Abstract:
The tripartite ATP-independent periplasmic (TRAP) transporters use an extra cytoplasmic substrate binding protein (SBP) to transport a wide variety of substrates in bacteria and archaea. The SBP can adopt an open- or closed state depending on the presence of substrate. The two transmembrane domains of TRAP transporters form a monomeric elevator whose function is strictly dependent on the presence of a sodium ion gradient. Insights from experimental structures, structural predictions and molecular modeling have suggested a conformational coupling between the membrane elevator and the substrate binding protein. Here, we use a disulfide engineering approach to lock the TRAP transporter HiSiaPQM from Haemophilus influenzae in different conformational states. The SBP, HiSiaP, is locked in its substrate-bound form and the transmembrane elevator, HiSiaQM, is locked in either its assumed inward- or outward-facing states. We characterize the disulfide-locked constructs and use single-molecule total internal reflection fluorescence (TIRF) microscopy to study their interactions. Our experiments demonstrate that the SBP and the transmembrane elevator are indeed conformationally coupled, meaning that the open and closed state of the SBP recognize specific conformational states of the transporter and vice versa.
Organizational Affiliation:
Institute of Structural Biology, University of Bonn, Venusberg-Campus 1, 53127, Bonn, Germany.