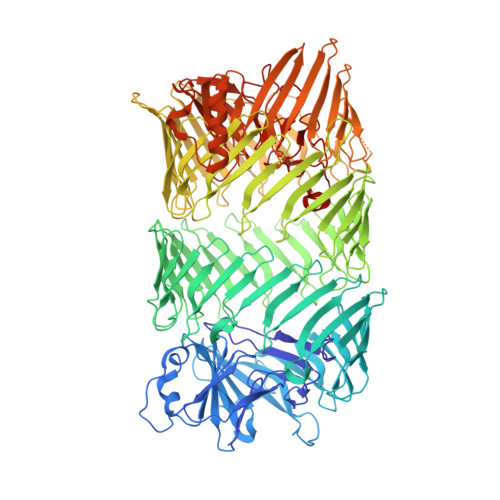
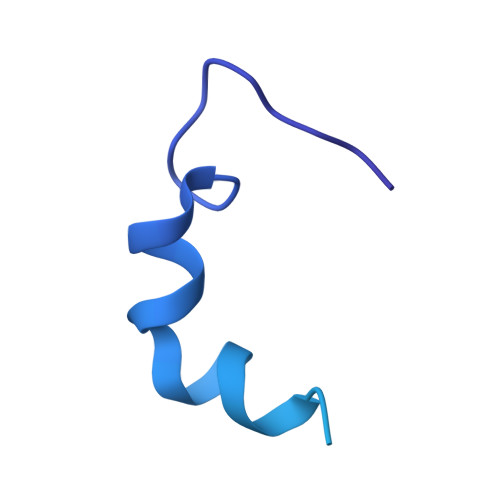
Structural and functional insights into the delivery of a bacterial Rhs pore-forming toxin to the membrane.
Gonzalez-Magana, A., Tascon, I., Altuna-Alvarez, J., Queralt-Martin, M., Colautti, J., Velazquez, C., Zabala, M., Rojas-Palomino, J., Cardenas, M., Alcaraz, A., Whitney, J.C., Ubarretxena-Belandia, I., Albesa-Jove, D.(2023) Nat Commun 14: 7808-7808
- PubMed: 38016939
- DOI: https://doi.org/10.1038/s41467-023-43585-5
- Primary Citation of Related Structures:
8CP6 - PubMed Abstract:
Bacterial competition is a significant driver of toxin polymorphism, which allows continual compensatory evolution between toxins and the resistance developed to overcome their activity. Bacterial Rearrangement hot spot (Rhs) proteins represent a widespread example of toxin polymorphism. Here, we present the 2.45 Å cryo-electron microscopy structure of Tse5, an Rhs protein central to Pseudomonas aeruginosa type VI secretion system-mediated bacterial competition. This structural insight, coupled with an extensive array of biophysical and genetic investigations, unravels the multifaceted functional mechanisms of Tse5. The data suggest that interfacial Tse5-membrane binding delivers its encapsulated pore-forming toxin fragment to the target bacterial membrane, where it assembles pores that cause cell depolarisation and, ultimately, bacterial death.
Organizational Affiliation:
Instituto Biofisika (CSIC, UPV/EHU), Fundación Biofísica Bizkaia/Biofisika Bizkaia Fundazioa (FBB), 48940, Leioa, Spain.