Structure of a hydrophobic leucinostatin derivative determined by host lattice display.
Kiss, C., Gall, F.M., Dreier, B., Adams, M., Riedl, R., Pluckthun, A., Mittl, P.R.E.(2022) Acta Crystallogr D Struct Biol 78: 1439-1450
- PubMed: 36458615
- DOI: https://doi.org/10.1107/S2059798322010762
- Primary Citation of Related Structures:
8A19, 8A1A - PubMed Abstract:
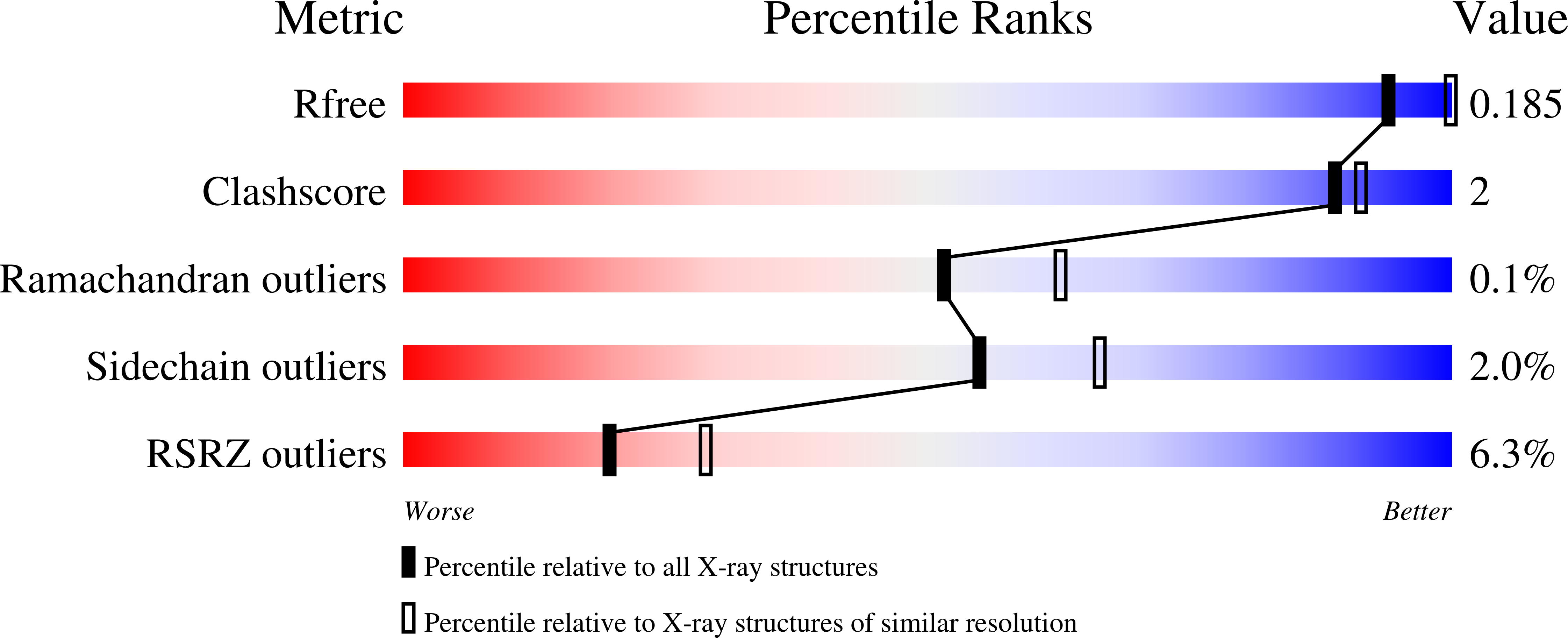
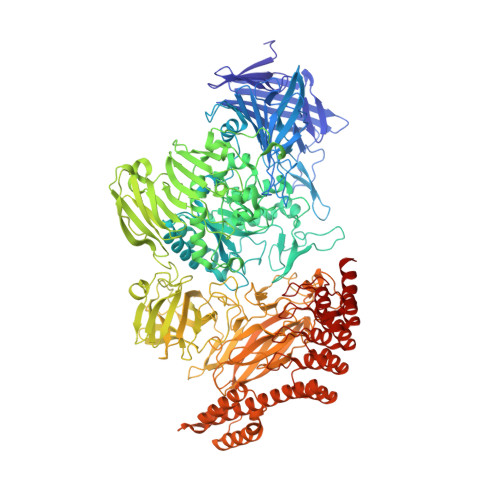
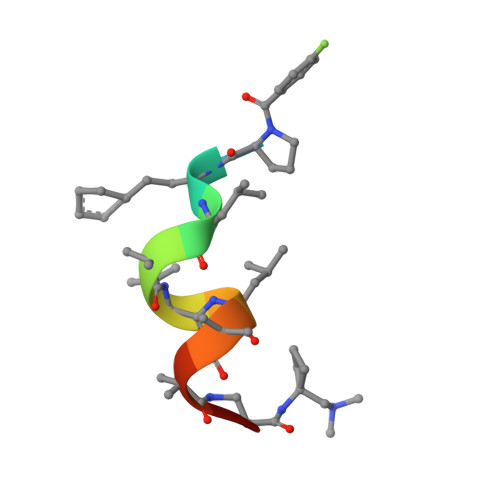
Peptides comprising many hydrophobic amino acids are almost insoluble under physiological buffer conditions, which complicates their structural analysis. To investigate the three-dimensional structure of the hydrophobic leucinostatin derivative ZHAWOC6027, the previously developed host lattice display technology was applied. Two designed ankyrin-repeat proteins (DARPins) recognizing a biotinylated ZHAWOC6027 derivative were selected from a diverse library by ribosome display under aqueous buffer conditions. ZHAWOC6027 was immobilized by means of the DARPin in the host lattice and the structure of the complex was determined by X-ray diffraction. ZHAWOC6027 adopts a distorted α-helical conformation. Comparison with the structures of related compounds that have been determined in organic solvents reveals elevated flexibility of the termini, which might be functionally important.
Organizational Affiliation:
Department of Biochemistry, University of Zürich, Winterthurerstrasse 190, 8057 Zürich, Switzerland.