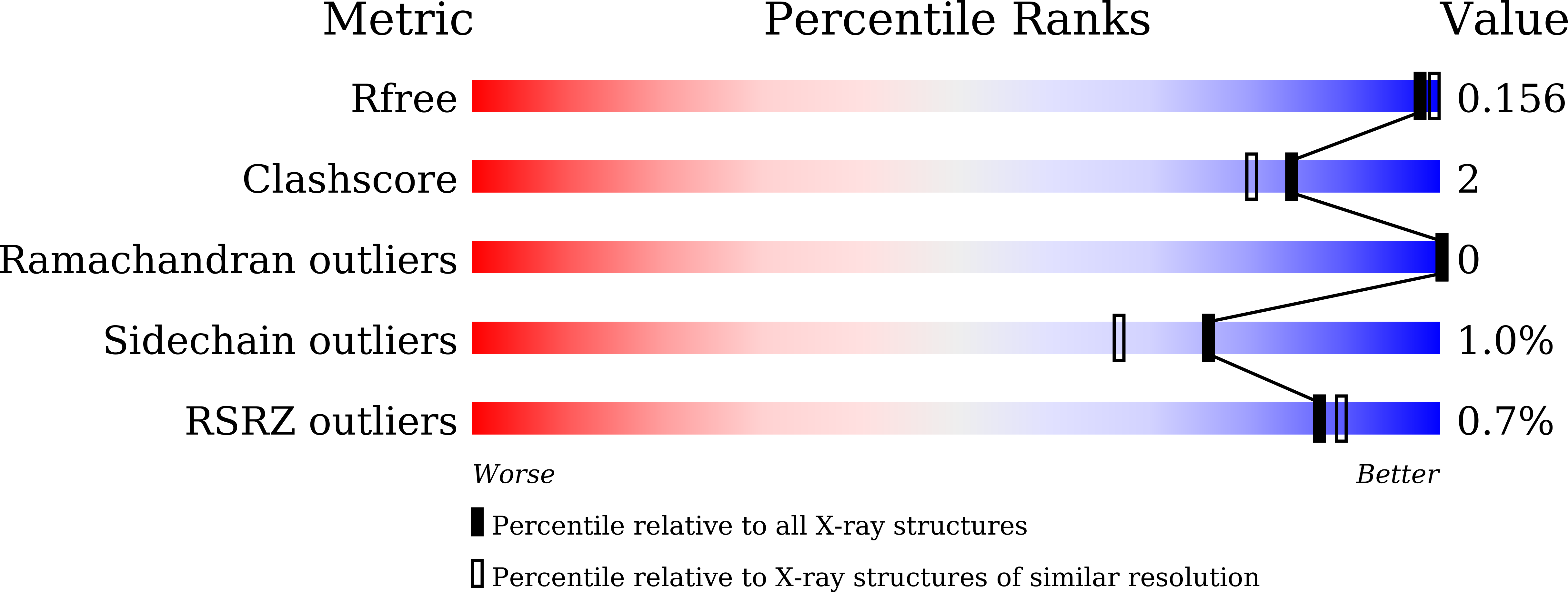
Structural analysis of Tris binding in beta-glucosidases.
Nam, K.H.(2024) Biochem Biophys Res Commun 700: 149608-149608
- PubMed: 38306932
- DOI: https://doi.org/10.1016/j.bbrc.2024.149608
- Primary Citation of Related Structures:
8XPC, 8XPD, 8XPE - PubMed Abstract:
β-glucosidases (Bgls) are glycosyl hydrolases that catalyze the conversion of cellobiose or glucosyl-polysaccharide into glucose. Bgls are widely used in industry to produce bioethanol, wine and juice, and feed. Tris (tris(hydroxymethyl)aminomethane) is an organic compound that can inhibit the hydrolase activity of some Bgls, but the inhibition state and selectivity have not been fully elucidated. Here, three crystal structures of Thermoanaerobacterium saccharolyticum Bgl complexed with the Tris molecule were determined at 1.55-1.95 Å. The configuration of Tris binding to TsaBgl remained consistent across three crystal structures, and the amino acids interacting with the Tris molecule were conserved across Bgl enzymes. The positions O1 and O3 atoms of Tris exhibit the same binding moiety as the hydroxyl group of the glucose molecule. Tris molecules are stably positioned at the glycone site and coordinate with surrounding water molecules. The Tris-binding configuration of TsaBgl is similar to that of HjeBgl, HgaBgl, ManBgl, and KflBgl, but the arrangement of the water molecule coordinating Tris at the aglycone site differs. Meanwhile, both the arrangement of Tris and the water molecules in ubBgl, NkoBgl, and SfrBgl differ from those in TsaBgl. The binding configuration and affinity of the Tris molecule for Bgl may be affected by the residues on the aglycone and gatekeeper regions. This result will extend our knowledge of the inhibitory effect of Tris molecules on TsaBgl.
Organizational Affiliation:
College of General Education, Kookmin University, Seoul, 20707, Republic of Korea. Electronic address: structure@kookmin.ac.kr.