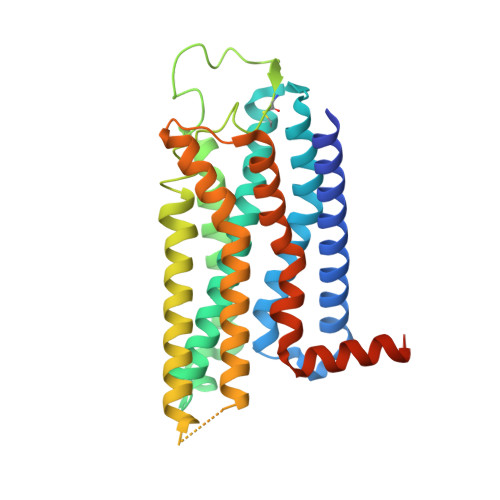
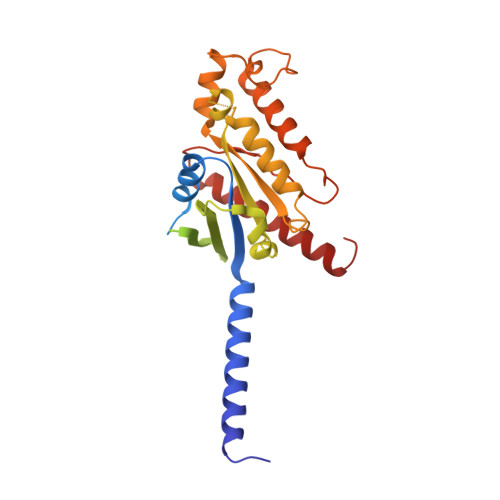
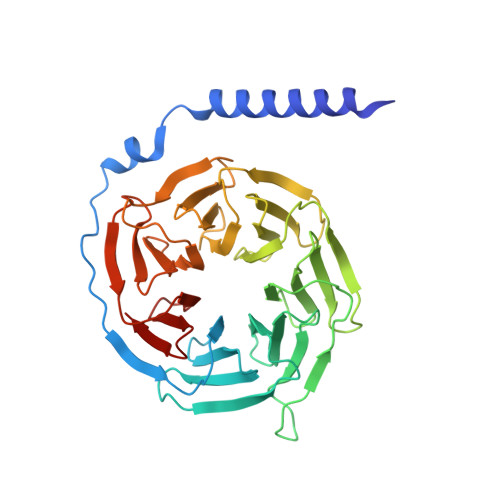
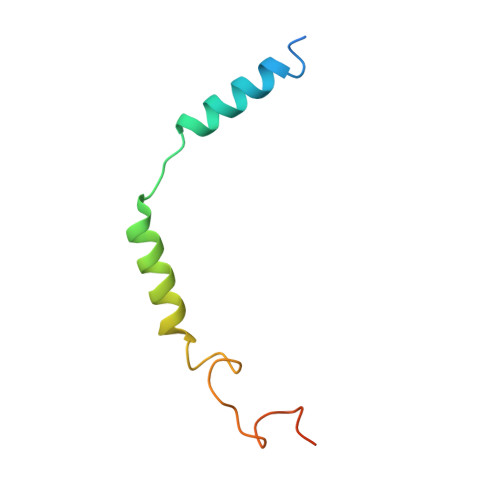
Cryo-EM structures of adenosine receptor A 3 AR bound to selective agonists.
Cai, H., Guo, S., Xu, Y., Sun, J., Li, J., Xia, Z., Jiang, Y., Xie, X., Xu, H.E.(2024) Nat Commun 15: 3252-3252
- PubMed: 38627384
- DOI: https://doi.org/10.1038/s41467-024-47207-6
- Primary Citation of Related Structures:
8X16, 8X17 - PubMed Abstract:
The adenosine A 3 receptor (A 3 AR), a key member of the G protein-coupled receptor family, is a promising therapeutic target for inflammatory and cancerous conditions. The selective A 3 AR agonists, CF101 and CF102, are clinically significant, yet their recognition mechanisms remained elusive. Here we report the cryogenic electron microscopy structures of the full-length human A 3 AR bound to CF101 and CF102 with heterotrimeric G i protein in complex at 3.3-3.2 Å resolution. These agonists reside in the orthosteric pocket, forming conserved interactions via their adenine moieties, while their 3-iodobenzyl groups exhibit distinct orientations. Functional assays reveal the critical role of extracellular loop 3 in A 3 AR's ligand selectivity and receptor activation. Key mutations, including His 3.37 , Ser 5.42 , and Ser 6.52 , in a unique sub-pocket of A 3 AR, significantly impact receptor activation. Comparative analysis with the inactive A 2A AR structure highlights a conserved receptor activation mechanism. Our findings provide comprehensive insights into the molecular recognition and signaling of A 3 AR, paving the way for designing subtype-selective adenosine receptor ligands.
Organizational Affiliation:
State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China. caihongmin@simm.ac.cn.