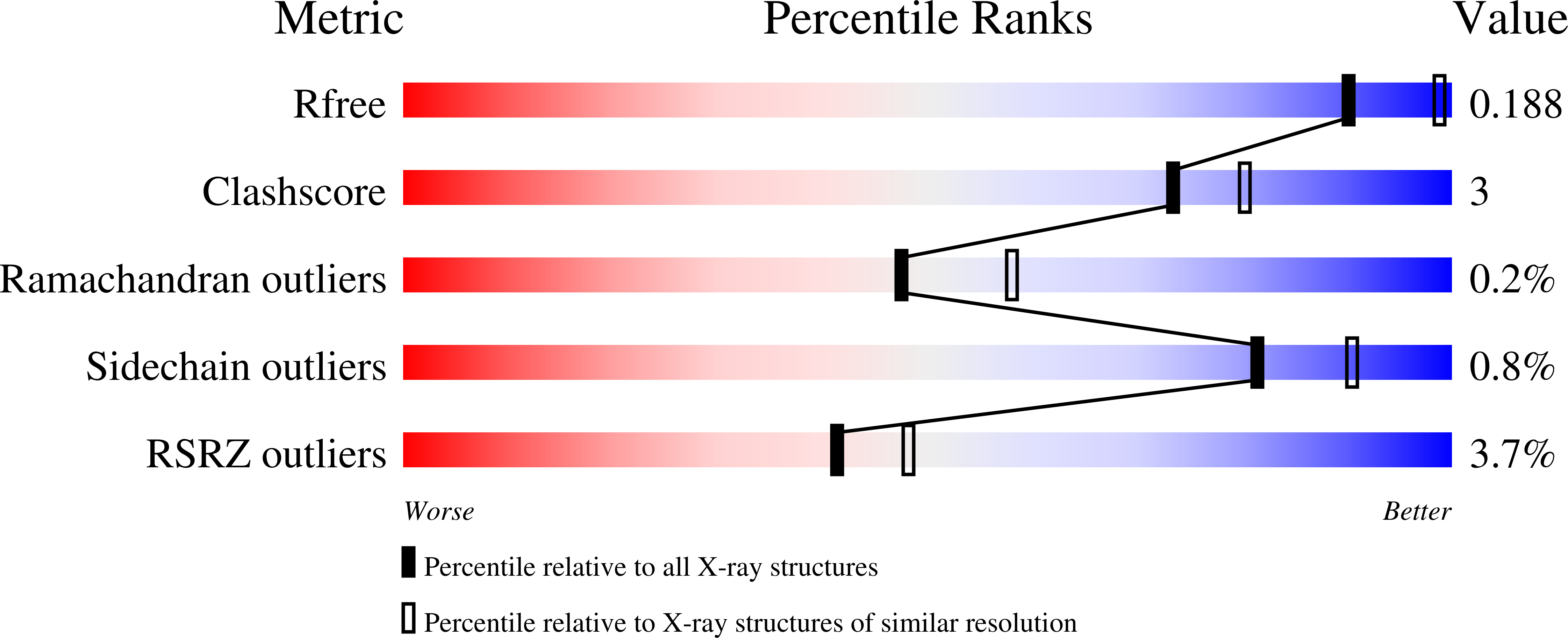
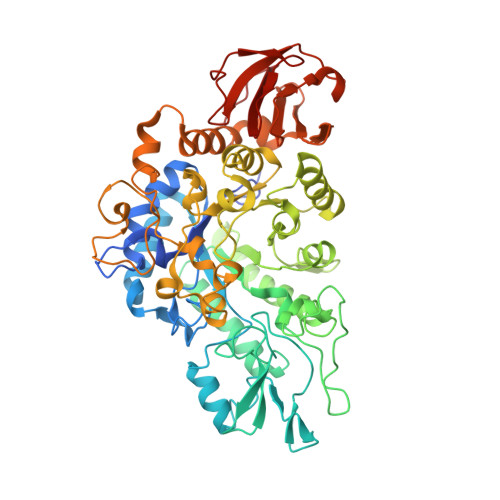
Structural and functional comparisons of salivary alpha-glucosidases from the mosquito vectors Aedes aegypti, Anopheles gambiae, and Culex quinquefasciatus.
Williams, A.E., Gittis, A.G., Botello, K., Cruz, P., Martin-Martin, I., Valenzuela Leon, P.C., Sumner, B., Bonilla, B., Calvo, E.(2024) Insect Biochem Mol Biol 167: 104097-104097
- PubMed: 38428508
- DOI: https://doi.org/10.1016/j.ibmb.2024.104097
- Primary Citation of Related Structures:
8SLV - PubMed Abstract:
Mosquito vectors of medical importance both blood and sugar feed, and their saliva contains bioactive molecules that aid in both processes. Although it has been shown that the salivary glands of several mosquito species exhibit α-glucosidase activities, the specific enzymes responsible for sugar digestion remain understudied. We therefore expressed and purified three recombinant salivary α-glucosidases from the mosquito vectors Aedes aegypti, Anopheles gambiae, and Culex quinquefasciatus and compared their functions and structures. We found that all three enzymes were expressed in the salivary glands of their respective vectors and were secreted into the saliva. The proteins, as well as mosquito salivary gland extracts, exhibited α-glucosidase activity, and the recombinant enzymes displayed preference for sucrose compared to p-nitrophenyl-α-D-glucopyranoside. Finally, we solved the crystal structure of the Ae. aegypti α-glucosidase bound to two calcium ions at a 2.3 Ångstrom resolution. Molecular docking suggested that the Ae. aegypti α-glucosidase preferred di- or polysaccharides compared to monosaccharides, consistent with enzymatic activity assays. Comparing structural models between the three species revealed a high degree of similarity, suggesting similar functional properties. We conclude that the α-glucosidases studied herein are important enzymes for sugar digestion in three mosquito species.
Organizational Affiliation:
Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD, 20852, USA.