Structural characterization of lytic transglycosylase MltD of Pseudomonas aeruginosa, a target for the natural product bulgecin A.
Miguel-Ruano, V., Feltzer, R., Batuecas, M.T., Ramachandran, B., El-Araby, A.M., Avila-Cobian, L.F., De Benedetti, S., Mobashery, S., Hermoso, J.A.(2024) Int J Biol Macromol 267: 131420-131420
- PubMed: 38583835
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.131420
- Primary Citation of Related Structures:
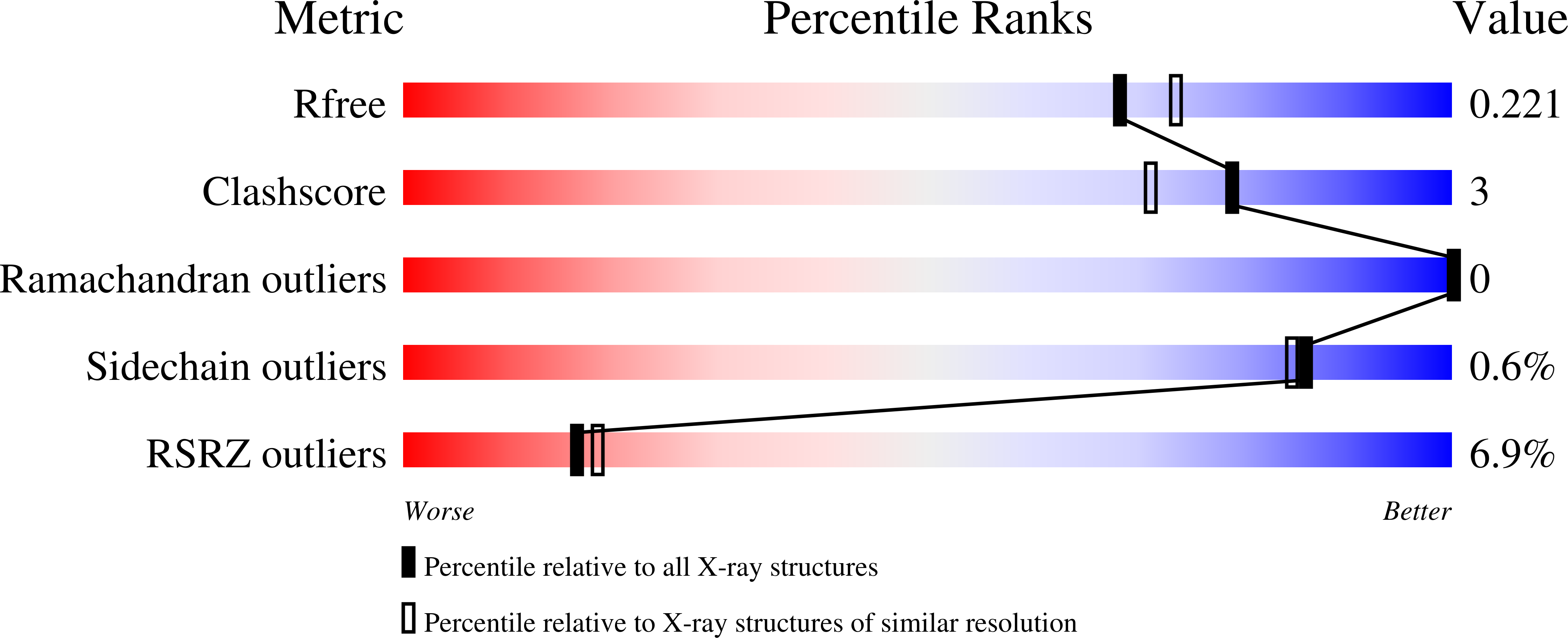
8RHE, 8RHF, 8RHI - PubMed Abstract:
Natural product bulgecin A potentiates the activity of β-lactam antibiotics by inhibition of three lytic transglycosylases in Pseudomonas aeruginosa, of which MltD is one. MltD exhibits both endolytic and exolytic reactions in the turnover of the cell-wall peptidoglycan and tolerates the presence or absence of stem peptides in its substrates. The present study reveals structural features of the multimodular MltD, presenting a catalytic module and four cell-wall-binding LysM modules that account for these attributes. Three X-ray structures are reported herein for MltD that disclose one unpredicted LysM module tightly attached to the catalytic domain, whereas the other LysM modules are mobile, and connected to the catalytic domain through long flexible linkers. The formation of crystals depended on the presence of bulgecin A. The expansive active-site cleft is highlighted by the insertion of a helical region, a hallmark of the family 1D of lytic transglycosylases, which was mapped out in a ternary complex of MltD:bulgecinA:chitotetraose, revealing at the minimum the presence of eight subsites (from -4 to +4, with the seat of reaction at subsites -1 and + 1) for binding of sugars of the substrate for the endolytic reaction. The mechanism of the exolytic reaction is revealed in one of the structures, showing how the substrate's terminal anhydro-NAM moiety could be sequestered at subsite +2. Our results provide the structural insight for both the endolytic and exolytic activities of MltD during cell-wall-turnover events.
Organizational Affiliation:
Department of Crystallography and Structural Biology, Instituto de Química-Física "Blas Cabrera", Consejo Superior de Investigaciones Científicas, Madrid, Spain.