Design, Synthesis, and Biological Evaluation of Trisubstituted Piperazine Derivatives as Noncovalent Severe Acute Respiratory Syndrome Coronavirus 2 Main Protease Inhibitors with Improved Antiviral Activity and Favorable Druggability.
Gao, S., Song, L., Sylvester, K., Mercorelli, B., Loregian, A., Toth, K., Weisse, R.H., Useini, A., Strater, N., Yang, M., Ye, B., Tollefson, A.E., Muller, C.E., Liu, X., Zhan, P.(2023) J Med Chem 66: 16426-16440
- PubMed: 37992202
- DOI: https://doi.org/10.1021/acs.jmedchem.3c01876
- Primary Citation of Related Structures:
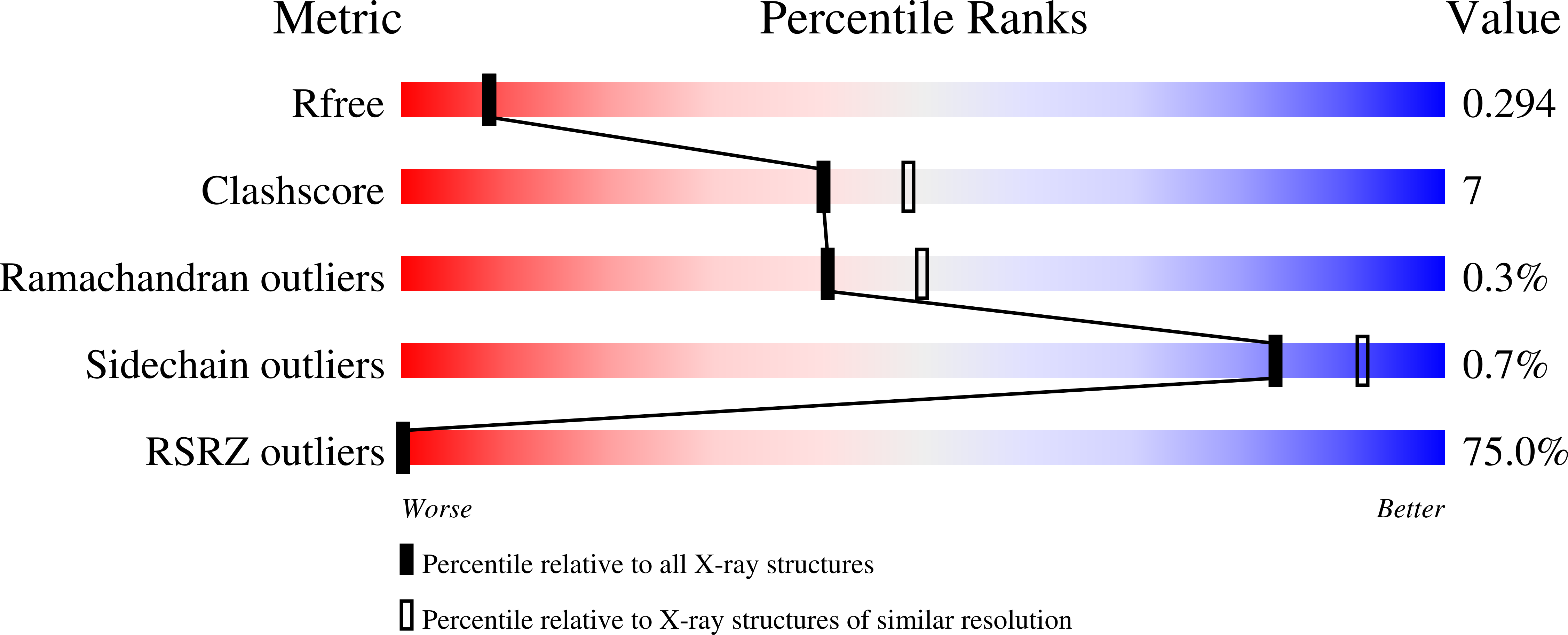
8Q71 - PubMed Abstract:
The ongoing transmission of SARS-CoV-2 necessitates the development of additional potent antiviral agents capable of combating the current highly infectious variants and future coronaviruses. Here, we present the discovery of potent nonpeptide main protease (M pro ) inhibitors with prominent antiviral activity and improved pharmacokinetic properties. Three series of 1,2,4-trisubstituted piperazine derivatives were designed and synthesized, and the optimal GC-78-HCl demonstrated high enzyme-inhibitory potency (IC 50 = 0.19 μM) and exhibited excellent antiviral activity (EC 50 = 0.40 μM), reaching the same level as Nirmatrelvir (EC 50 = 0.38 μM). Additionally, GC-78-HCl displayed potent antiviral activities against various SARS-CoV-2 variants as well as HCoV-OC43 and HCoV-229E, indicating its potential broad-spectrum anticoronaviral activity. Notably, the pharmacokinetic properties of GC-78-HCl were somewhat enhanced compared to those of the lead compound. Furthermore, the cocrystal and molecular docking elucidated the mechanism of action. In conclusion, we discovered a novel nonpeptidic M pro inhibitor with promising antiviral activity and a favorable pharmacokinetic profile.
Organizational Affiliation:
Department of Medicinal Chemistry, Key Laboratory of Chemical Biology, Ministry of Education, School of Pharmaceutical Sciences, Shandong University, Ji'nan 250012, China.