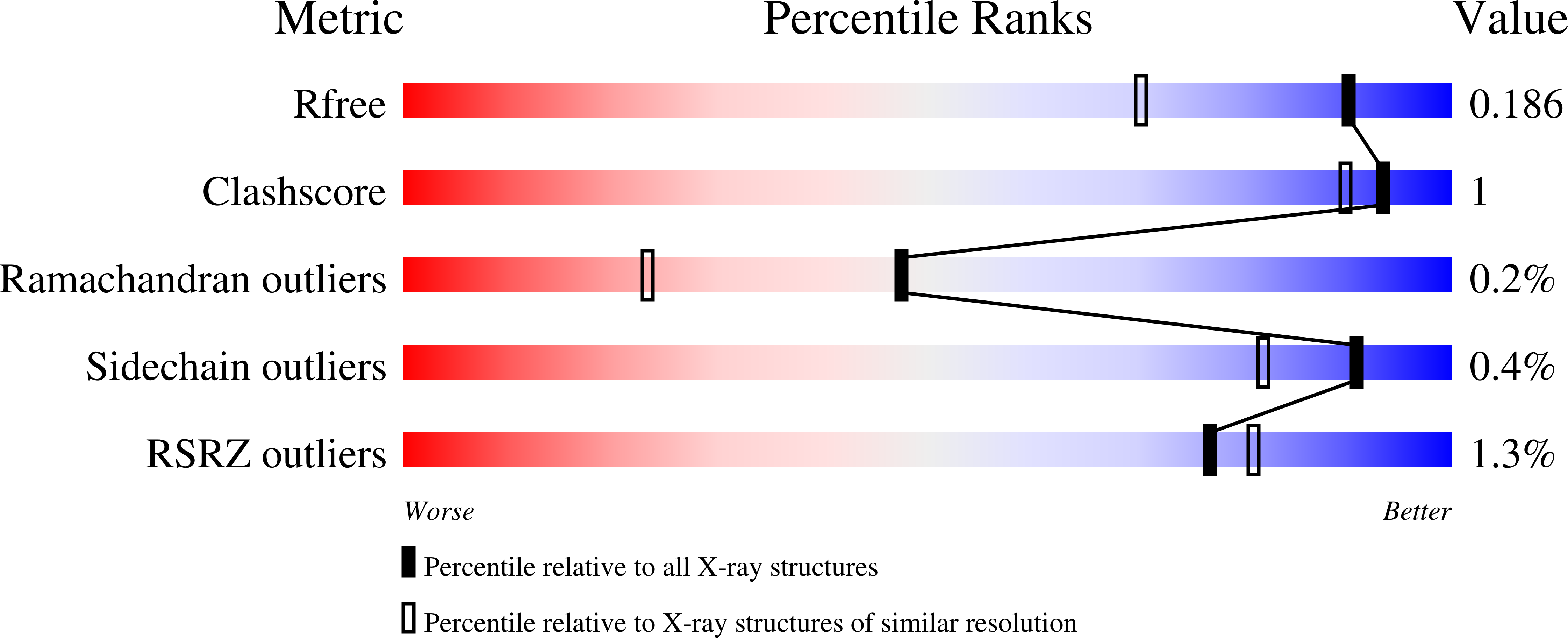
The development of a broad-spectrum retaining beta-exo-galactosidase activity-based probe.
Kuo, C.L., Su, Q., van den Nieuwendijk, A.M.C.H., Beenakker, T.J.M., Offen, W.A., Willems, L.I., Boot, R.G., Sarris, A.J., Marques, A.R.A., Codee, J.D.C., van der Marel, G.A., Florea, B.I., Davies, G.J., Overkleeft, H.S., Aerts, J.M.F.G.(2023) Org Biomol Chem 21: 7813-7820
- PubMed: 37724332
- DOI: https://doi.org/10.1039/d3ob01261a
- Primary Citation of Related Structures:
8PEJ - PubMed Abstract:
Acid β-galactosidase (GLB1) and galactocerebrosidase (GALC) are retaining exo-β-galactosidases involved in lysosomal glycoconjugate metabolism. Deficiency of GLB1 may result in the lysosomal storage disorders GM1 gangliosidosis, Morquio B syndrome, and galactosialidosis, and deficiency of GALC may result in Krabbe disease. Activity-based protein profiling (ABPP) is a powerful technique to assess the activity of retaining glycosidases in relation to health and disease. This work describes the use of fluorescent and biotin-carrying activity-based probes (ABPs) to assess the activity of both GLB1 and GALC in cell lysates, culture media, and tissue extracts. The reported ABPs, which complement the growing list of retaining glycosidase ABPs based on configurational isomers of cyclophellitol, should assist in fundamental and clinical research on various β-galactosidases, whose inherited deficiencies cause debilitating lysosomal storage disorders.
Organizational Affiliation:
Department of Medical Biochemistry, Leiden Institute of Chemistry, Leiden University, P. O. Box 9502, 2300 RA Leiden, The Netherlands. j.m.f.g.aerts@lic.leidenuniv.nl.