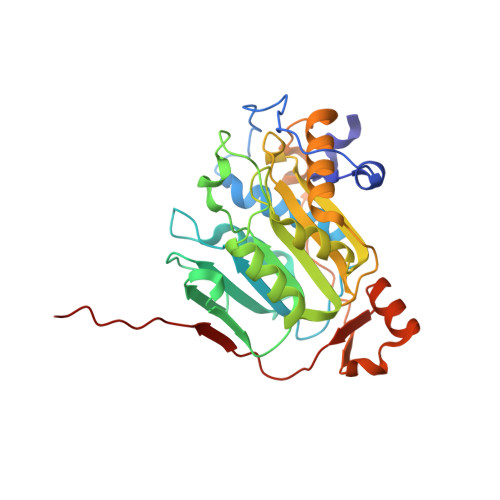
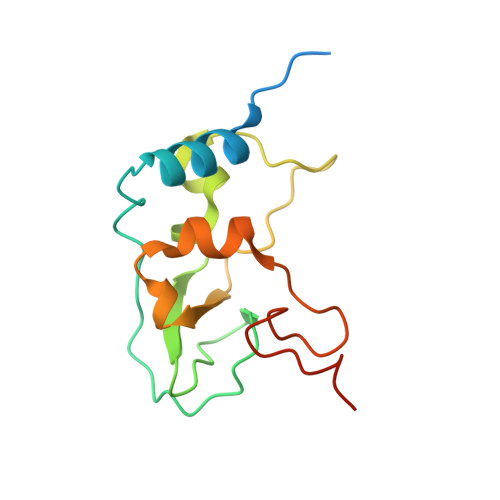
Crystal structures of Tubercidin and Adenosine bound to the active site of the SARS-CoV-2 methyltransferase nsp10-16
Kremling, V., Oberthuer, D., Yefanov, O., Galchenkova, M., Middendorf, P., Fernandez Garcia, Y., Ehrt, C., Falke, S., Kiene, A., Klopprogge, B., Chapman, H., Sprenger, J.To be published.