Discovery of 4,5,6,7-Tetrahydropyrazolo[1.5-a]pyrizine Derivatives as Core Protein Allosteric Modulators (CpAMs) for the Inhibition of Hepatitis B Virus.
Kou, B., Zhang, Z., Han, X., Zhou, Z., Xu, Z., Zhou, X., Shen, F., Zhou, Y., Tian, X., Yang, G., Young, J.A.T., Qiu, H., Ottaviani, G., Mayweg, A., Zhu, W., Shen, H.C., Liu, H., Hu, T.(2023) J Med Chem 66: 14116-14132
- PubMed: 37801325
- DOI: https://doi.org/10.1021/acs.jmedchem.3c01145
- Primary Citation of Related Structures:
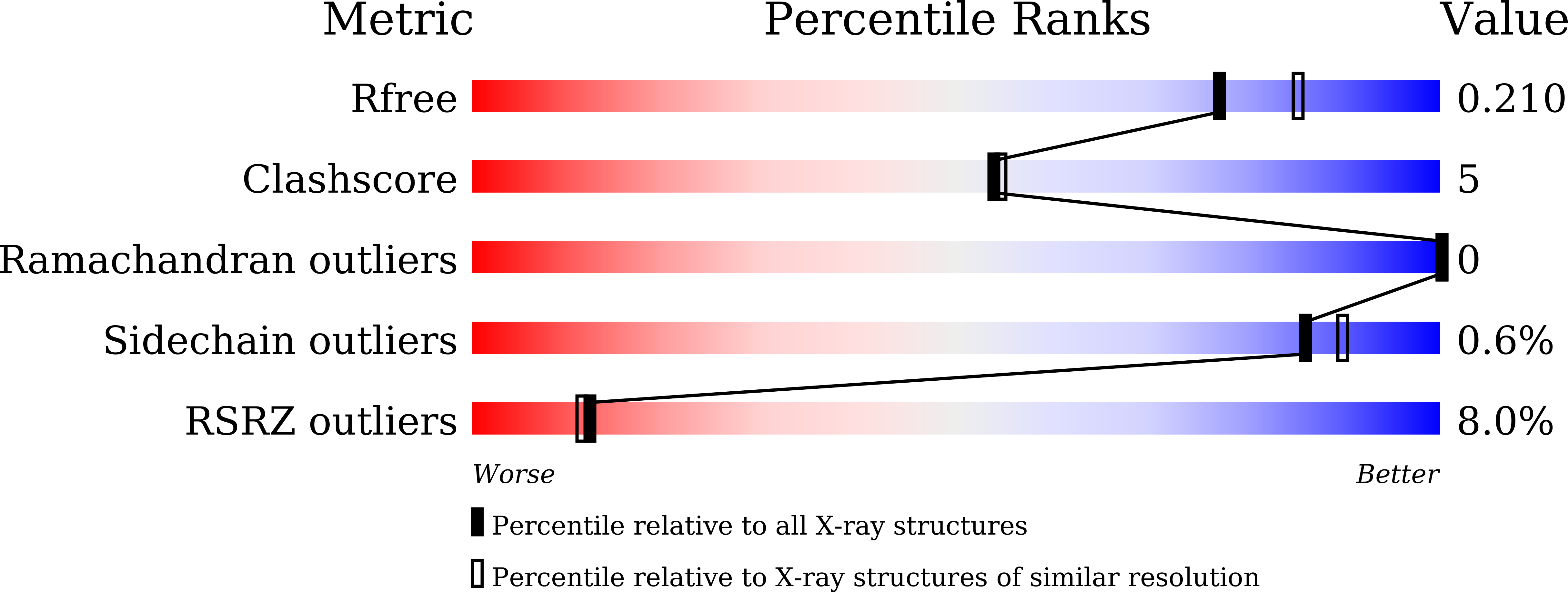
8KHU - PubMed Abstract:
Hepatitis B Virus (HBV) core protein allosteric modulators (CpAMs) are an attractive class of potential anti-HBV therapeutic agents. Here we describe the efforts toward the discovery of a series of 4,5,6,7-tetrahydropyrazolo[1,5- a ]pyrazine (THPP) compounds as HBV CpAMs that effectively inhibit a broad range of nucleos(t)ide-resistant HBV variants. The lead compound 45 demonstrated inhibition of HBV DNA viral load in a HBV AAV mouse model by oral administration.
Organizational Affiliation:
China Innovation Center of Roche, Building 5, 371 Lishizhen Road, Shanghai 201203, China.