Bifidobacterial GH146 beta-L-arabinofuranosidase for the removal of beta 1,3-L-arabinofuranosides on plant glycans.
Fujita, K., Tsunomachi, H., Lixia, P., Maruyama, S., Miyake, M., Dakeshita, A., Kitahara, K., Tanaka, K., Ito, Y., Ishiwata, A., Fushinobu, S.(2024) Appl Microbiol Biotechnol 108: 199-199
- PubMed: 38324037
- DOI: https://doi.org/10.1007/s00253-024-13014-8
- Primary Citation of Related Structures:
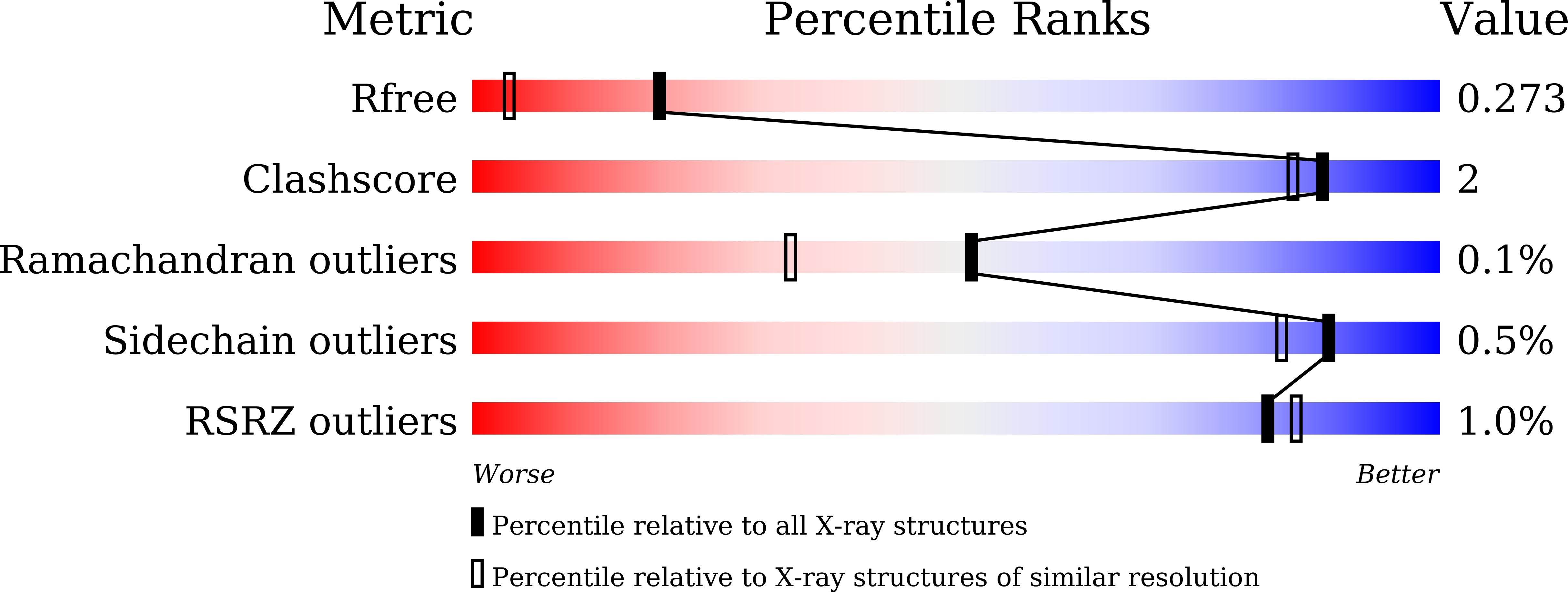
8K7X, 8K7Y - PubMed Abstract:
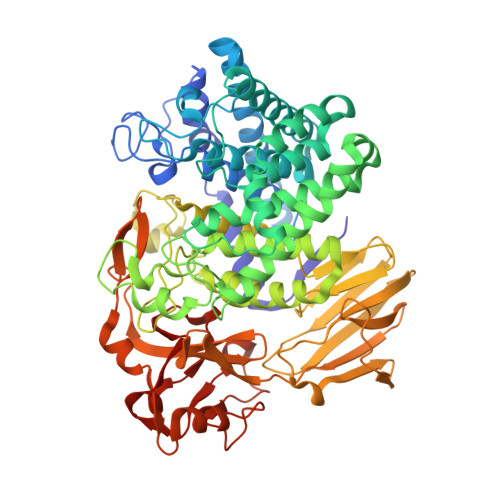
L-Arabinofuranosides with β-linkages are present in several plant molecules, such as arabinogalactan proteins (AGPs), extensin, arabinan, and rhamnogalacturonan-II. We previously characterized a β-L-arabinofuranosidase from Bifidobacterium longum subsp. longum JCM 1217, Bll1HypBA1, which was found to belong to the glycoside hydrolase (GH) family 127. This strain encodes two GH127 genes and two GH146 genes. In the present study, we characterized a GH146 β-L-arabinofuranosidase, Bll3HypBA1 (BLLJ_1848), which was found to constitute a gene cluster with AGP-degrading enzymes. This recombinant enzyme degraded AGPs and arabinan, which contain Araf-β1,3-Araf structures. In addition, the recombinant enzyme hydrolyzed oligosaccharides containing Araf-β1,3-Araf structures but not those containing Araf-β1,2-Araf and Araf-β1,5-Araf structures. The crystal structures of Bll3HypBA1 were determined at resolutions up to 1.7 Å. The monomeric structure of Bll3HypBA1 comprised a catalytic (α/α) 6 barrel and two β-sandwich domains. A hairpin structure with two β-strands was observed in Bll3HypBA1, to extend from a β-sandwich domain and partially cover the active site. The active site contains a Zn 2+ ion coordinated by Cys 3 -Glu and exhibits structural conservation of the GH127 cysteine glycosidase Bll1HypBA1. This is the first study to report on a β1,3-specific β-L-arabinofuranosidase. KEY POINTS: • β1,3-l-Arabinofuranose residues are present in arabinogalactan proteins and arabinans as a terminal sugar. • β-l-Arabinofuranosidases are widely present in intestinal bacteria. • Bll3HypBA1 is the first enzyme characterized as a β1,3-linkage-specific β-l-arabinofuranosidase.
Organizational Affiliation:
Faculty of Agriculture, Kagoshima University, 1-21-24 Korimoto, Kagoshima, Kagoshima, 890-0065, Japan. k4022897@kadai.jp.