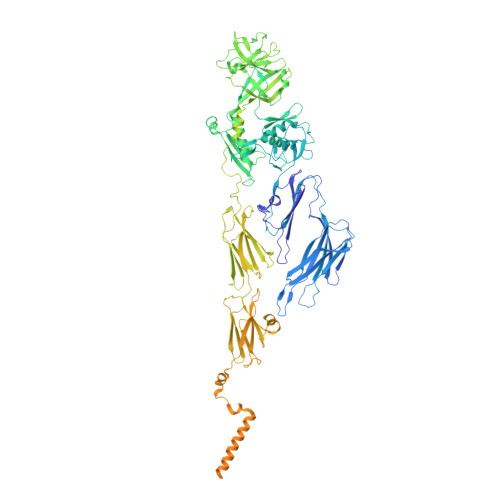
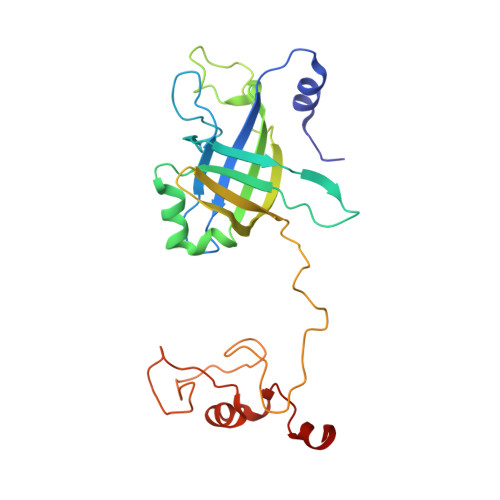
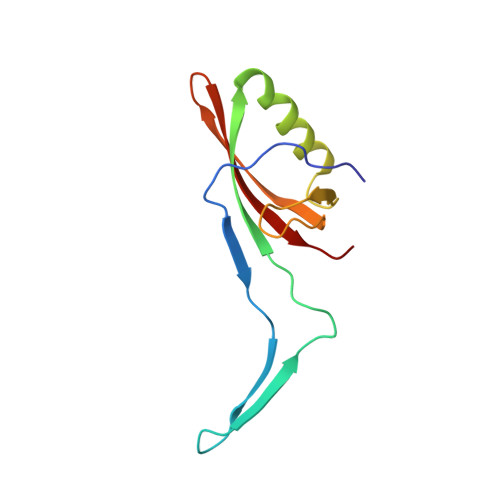
Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Xiao, H., Tan, L., Tan, Z., Zhang, Y., Chen, W., Li, X., Song, J., Cheng, L., Liu, H.(2023) PLoS Biol 21: e3002441-e3002441
- PubMed: 38096144
- DOI: https://doi.org/10.1371/journal.pbio.3002441
- Primary Citation of Related Structures:
8K35, 8K36, 8K37, 8K38, 8K39 - PubMed Abstract:
Siphophages have a long, flexible, and noncontractile tail that connects to the capsid through a neck. The phage tail is essential for host cell recognition and virus-host cell interactions; moreover, it serves as a channel for genome delivery during infection. However, the in situ high-resolution structure of the neck-tail complex of siphophages remains unknown. Here, we present the structure of the siphophage lambda "wild type," the most widely used, laboratory-adapted fiberless mutant. The neck-tail complex comprises a channel formed by stacked 12-fold and hexameric rings and a 3-fold symmetrical tip. The interactions among DNA and a total of 246 tail protein molecules forming the tail and neck have been characterized. Structural comparisons of the tail tips, the most diversified region across the lambda and other long-tailed phages or tail-like machines, suggest that their tail tip contains conserved domains, which facilitate tail assembly, receptor binding, cell adsorption, and DNA retaining/releasing. These domains are distributed in different tail tip proteins in different phages or tail-like machines. The side tail fibers are not required for the phage particle to orient itself vertically to the surface of the host cell during attachment.
Organizational Affiliation:
Institute of Interdisciplinary Studies, Key Laboratory for Matter Microstructure and Function of Hunan Province, Key Laboratory of Low-dimensional Quantum Structures and Quantum Control, School of Physics and Electronics, Hunan Normal University, Changsha, China.