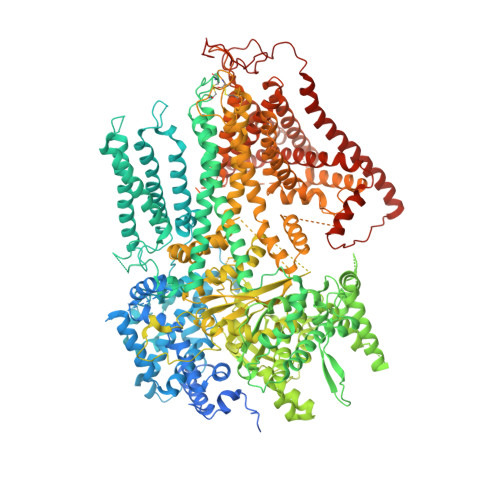
Structure of a fungal 1,3-beta-glucan synthase.
Zhao, C.R., You, Z.L., Chen, D.D., Hang, J., Wang, Z.B., Ji, M., Wang, L.X., Zhao, P., Qiao, J., Yun, C.H., Bai, L.(2023) Sci Adv 9: eadh7820-eadh7820
- PubMed: 37703377
- DOI: https://doi.org/10.1126/sciadv.adh7820
- Primary Citation of Related Structures:
8JZN - PubMed Abstract:
1,3-β-Glucan serves as the primary component of the fungal cell wall and is produced by 1,3-β-glucan synthase located in the plasma membrane. This synthase is a molecular target for antifungal drugs such as echinocandins and the triterpenoid ibrexafungerp. In this study, we present the cryo-electron microscopy structure of Saccharomyces cerevisiae 1,3-β-glucan synthase (Fks1) at 2.47-Å resolution. The structure reveals a central catalytic region adopting a cellulose synthase fold with a cytosolic conserved GT-A-type glycosyltransferase domain and a closed transmembrane channel responsible for glucan transportation. Two extracellular disulfide bonds are found to be crucial for Fks1 enzymatic activity. Through structural comparative analysis with cellulose synthases and structure-guided mutagenesis studies, we gain previously unknown insights into the molecular mechanisms of fungal 1,3-β-glucan synthase.
Organizational Affiliation:
Department of Biochemistry and Biophysics, School of Basic Medical Sciences, Peking University, Beijing, China.