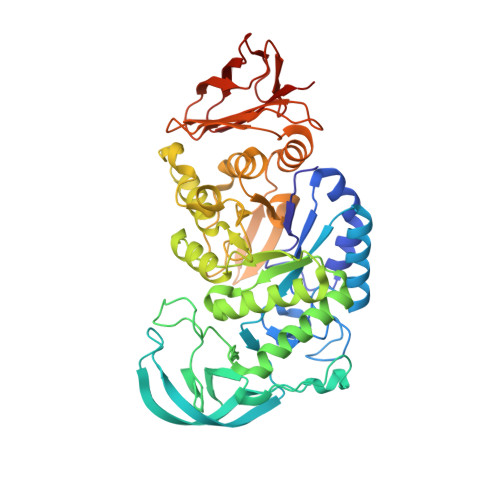
The novel amylase function of the carboxyl terminal domain of Amy63.
Sun, Y., Liu, G., Liu, G., Tang, H., Sun, C., Zhang, W., Chen, L.(2023) Biochem Biophys Res Commun 671: 10-17
- PubMed: 37290279
- DOI: https://doi.org/10.1016/j.bbrc.2023.05.071
- Primary Citation of Related Structures:
8JJM - PubMed Abstract:
α-amylase plays a crucial role in regulating metabolism and health by hydrolyzing of starch and glycogen. Despite comprehensive studies of this classic enzyme spanning over a century, the function of its carboxyl terminal domain (CTD) with a conserved eight β-strands is still not fully understood. Amy63, identified from a marine bacterium, was reported as a novel multifunctional enzyme with amylase, agarase and carrageenase activities. In this study, the crystal structure of Amy63 was determined at 1.8 Å resolution, revealing high conservation with some other amylases. Interestingly, the independent amylase activity of the carboxyl terminal domain of Amy63 (Amy63_CTD) was newly discovered by the plate-based assay and mass spectrometry. To date, the Amy63_CTD alone could be regarded as the smallest amylase subunit. Moreover, the significant amylase activity of Amy63_CTD was measured over a wide range of temperature and pH, with optimal activity at 60 °C and pH 7.5. The Small-angle X-ray scattering (SAXS) data showed that the high-order oligomeric assembly gradually formed with increasing concentration of Amy63_CTD, implying the novel catalytic mechanism as revealed by the assembly structure. Therefore, the discovery of the novel independent amylase activity of Amy63_CTD suggests a possible missing step or a new perspective in the complex catalytic process of Amy63 and other related α-amylases. This work may shed light on the design of nanozymes to process marine polysaccharides efficiently.
Organizational Affiliation:
Department of Medical Microbiology, Key Laboratory of Medical Molecular Virology of Ministries of Education and Health, School of Basic Medical Sciences, Fudan University, Shanghai, 200032, China; Fudan University Pudong Medical Center, Institutes of Biomedical Sciences, Fudan University, Shanghai, 200032, China; The Department of Systems Biology for Medicine, School of Basic Medical Sciences, Fudan University, Shanghai, 200032, China.