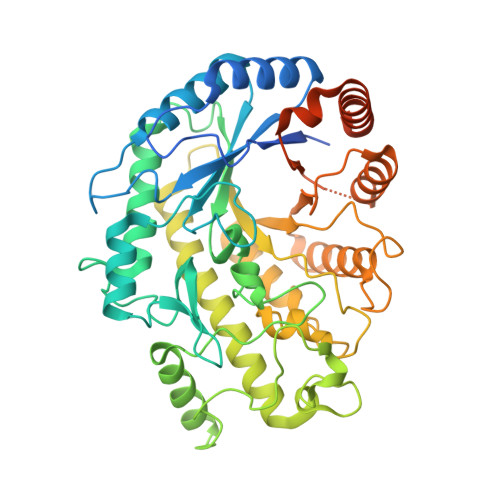
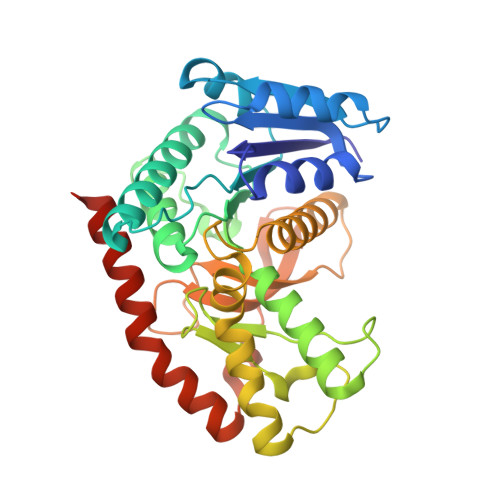
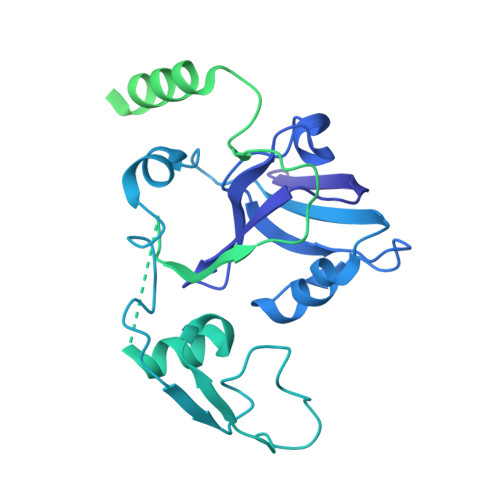
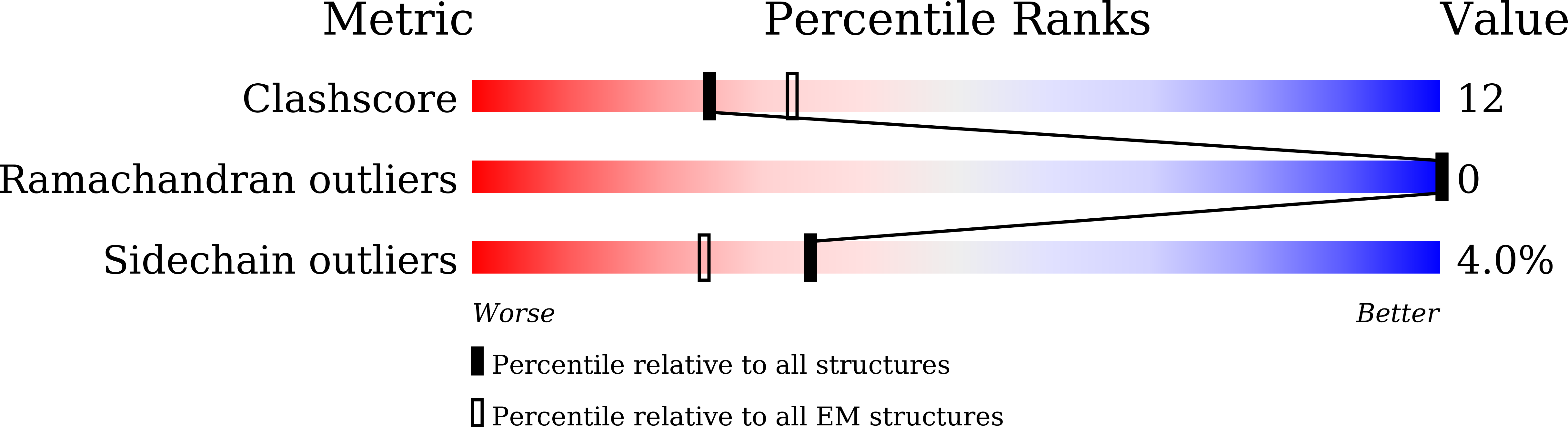The LIKE SEX FOUR 1-malate dehydrogenase complex functions as a scaffold to recruit beta-amylase to promote starch degradation.
Liu, J., Wang, X., Guan, Z., Wu, M., Wang, X., Fan, R., Zhang, F., Yan, J., Liu, Y., Zhang, D., Yin, P., Yan, J.(2023) Plant Cell 36: 194-212
- PubMed: 37804098
- DOI: https://doi.org/10.1093/plcell/koad259
- Primary Citation of Related Structures:
8J5D - PubMed Abstract:
In plant leaves, starch is composed of glucan polymers that accumulate in chloroplasts as the products of photosynthesis during the day; starch is mobilized at night to continuously provide sugars to sustain plant growth and development. Efficient starch degradation requires the involvement of several enzymes, including β-amylase and glucan phosphatase. However, how these enzymes cooperate remains largely unclear. Here, we show that the glucan phosphatase LIKE SEX FOUR 1 (LSF1) interacts with plastid NAD-dependent malate dehydrogenase (MDH) to recruit β-amylase (BAM1), thus reconstituting the BAM1-LSF1-MDH complex. The starch hydrolysis activity of BAM1 drastically increased in the presence of LSF1-MDH in vitro. We determined the structure of the BAM1-LSF1-MDH complex by a combination of cryo-electron microscopy, crosslinking mass spectrometry, and molecular docking. The starch-binding domain of the dual-specificity phosphatase and carbohydrate-binding module of LSF1 was docked in proximity to BAM1, thus facilitating BAM1 access to and hydrolysis of the polyglucans of starch, thus revealing the molecular mechanism by which the LSF1-MDH complex improves the starch degradation activity of BAM1. Moreover, LSF1 is phosphatase inactive, and the enzymatic activity of MDH was dispensable for starch degradation, suggesting nonenzymatic scaffold functions for LSF1-MDH in starch degradation. These findings provide important insights into the precise regulation of starch degradation.
Organizational Affiliation:
National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan 430070, China.