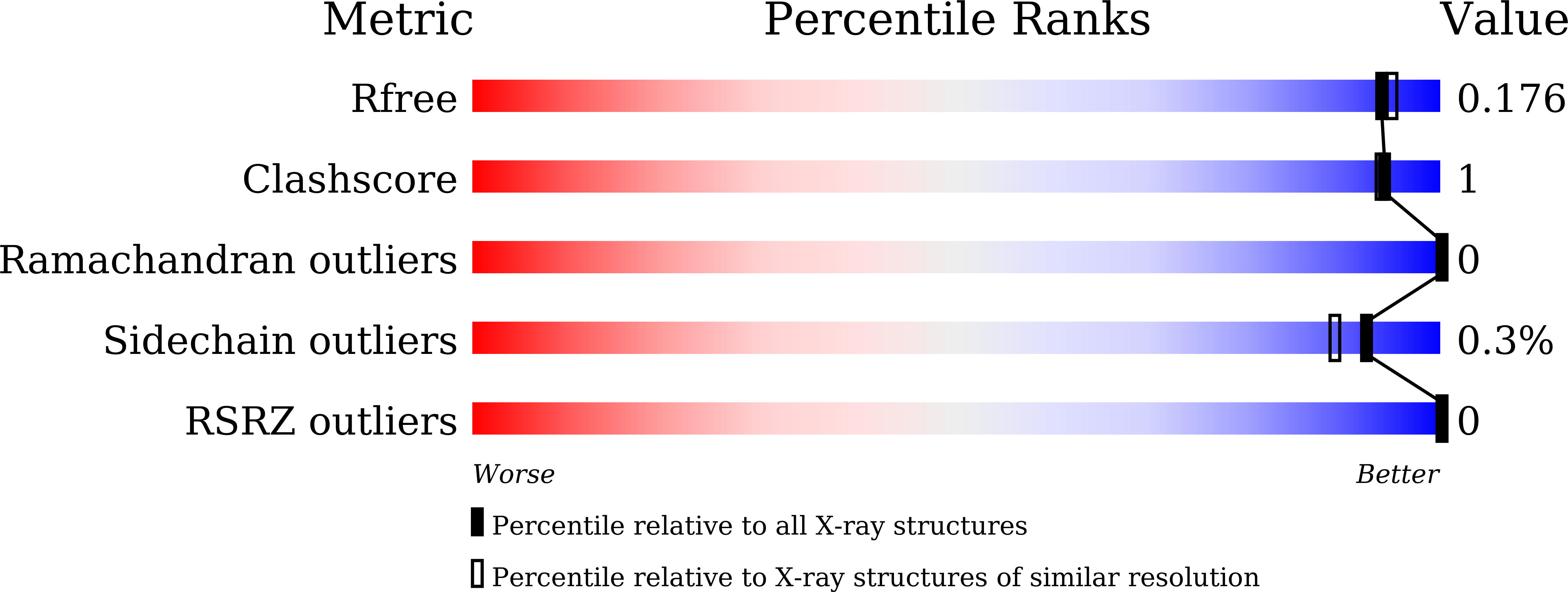
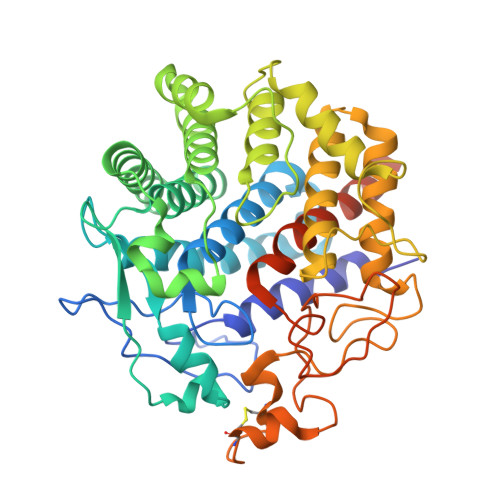
A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Kuroki, C., Hirano, Y., Nakazawa, M., Sakamoto, T., Tamada, T., Ueda, M.(2023) Curr Res Biotechnol 5