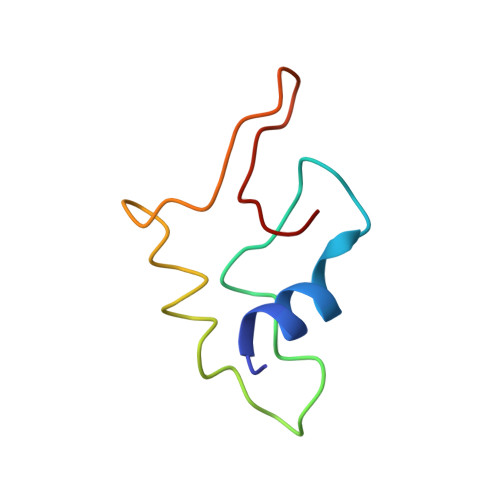
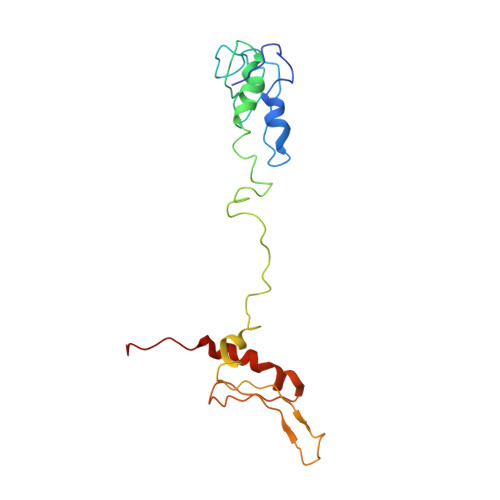
Structural study of novel vaccinia virus E3L and dsRNA-dependent protein kinase complex.
Kim, H.J., Han, C.W., Jeong, M.S., Jang, S.B.(2023) Biochem Biophys Res Commun 665: 1-9
- PubMed: 37146409
- DOI: https://doi.org/10.1016/j.bbrc.2023.04.107
- Primary Citation of Related Structures:
8I9J - PubMed Abstract:
E3L (RNA-binding protein E3) is one of the key IFN resistance genes encoded by VV and consists of 190 amino acids with a highly conserved carboxy-terminal double-stranded RNA-binding domain (dsRBD). PKR (dsRNA-dependent protein kinase) is an IFN-induced protein involved in anti-cell and antiviral activity. PKR inhibits the initiation of translation through alpha subunit of the initiation factor eIF2 (eIF2α) and mediates several transcription factors such as NF-κB, p53 or STATs. Activated PKR also induces apoptosis in vaccinia virus infection. E3L is required for viral IFN resistance and directly binds to PKR to block activation of PKR. In this work, we determined the three-dimensional complex structure of E3L and PKR using cryo-EM and determined the important residues involved in the interaction. In addition, PKR peptide binds to E3L and can increase protein levels of phosphorus-PKR and phosphorus-eIF2α-induced cell apoptosis through upregulation of phosphorus-PKR in HEK293 cells. Taken together, structural insights into E3L and PKR will provide a new optimization and development of vaccinia virus drugs.
Organizational Affiliation:
Department of Molecular Biology, College of Natural Sciences, Pusan National University, 2, Busandaehak-ro 63beon-gil, Geumjeong-gu, Busan, 46241, Republic of Korea.