The cryo-EM structure of cellobiose phosphorylase from Clostridium thermocellum in complex with cellobiose
Iriya, S., Kuga, T., Sunagawa, N., Igarashi, K.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cellobiose phosphorylase | 817 | Acetivibrio thermocellus | Mutation(s): 0 Gene Names: cbp EC: 2.4.1.20 | 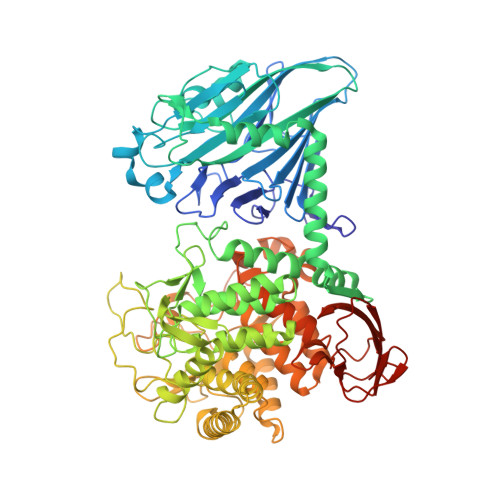 | |
UniProt | |||||
Find proteins for Q8VP44 (Acetivibrio thermocellus) Explore Q8VP44 Go to UniProtKB: Q8VP44 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8VP44 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900005 Query on PRD_900005 | C, D | beta-cellobiose | Oligosaccharide / Metabolism |  | |
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 3.3.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | 22J12566 |