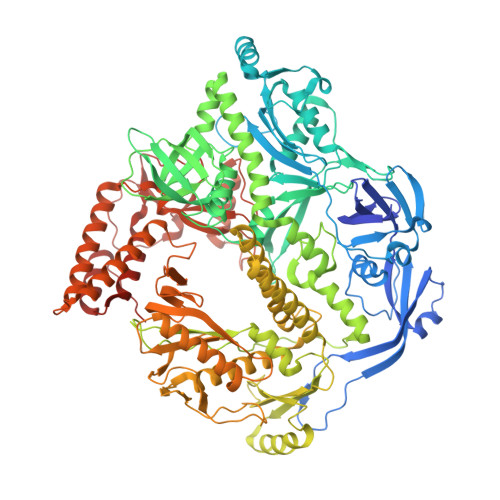
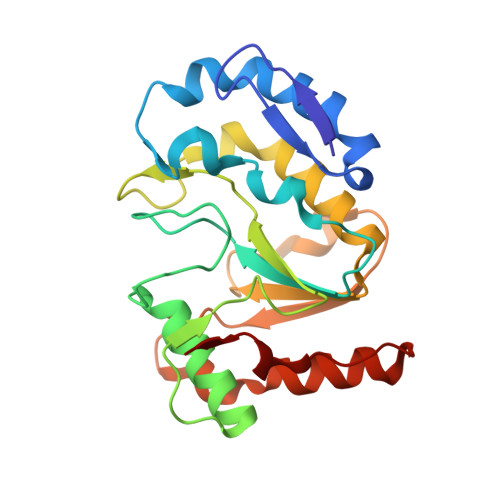
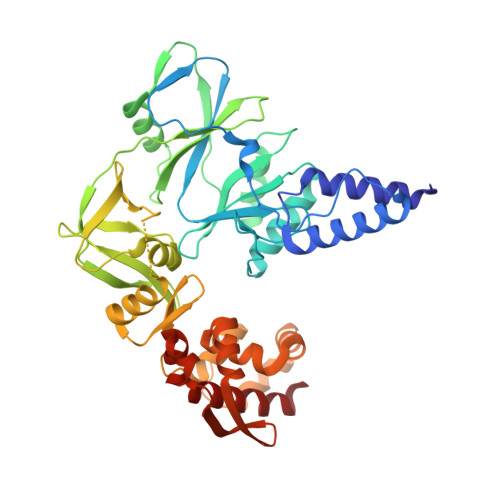
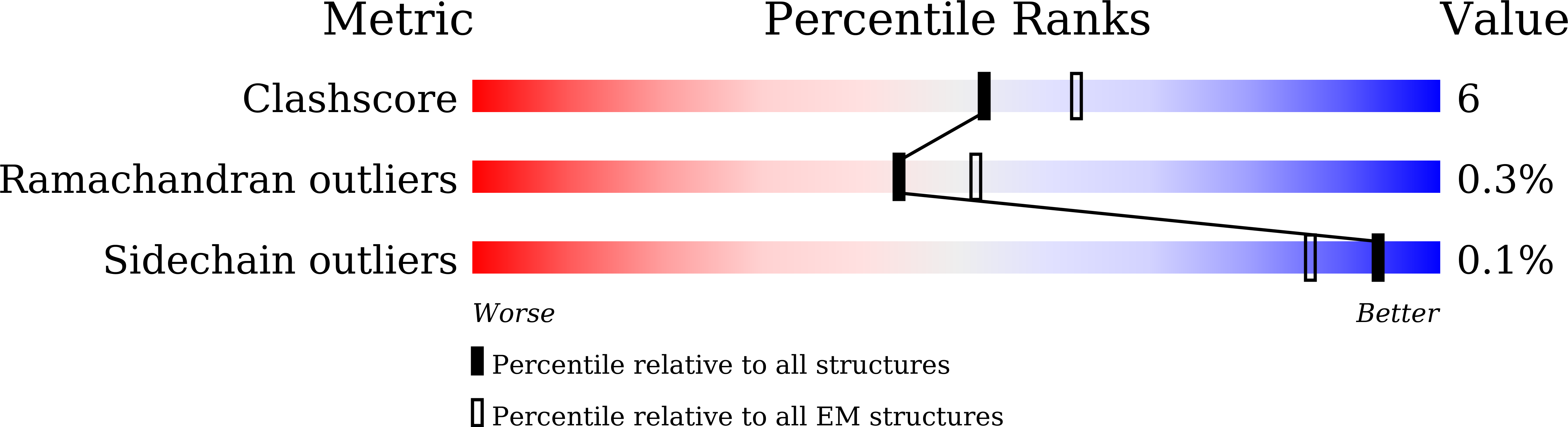Structure of monkeypox virus DNA polymerase holoenzyme.
Peng, Q., Xie, Y., Kuai, L., Wang, H., Qi, J., Gao, G.F., Shi, Y.(2023) Science 379: 100-105
- PubMed: 36520947
- DOI: https://doi.org/10.1126/science.ade6360
- Primary Citation of Related Structures:
8HG1 - PubMed Abstract:
The World Health Organization declared mpox (or monkeypox) a public health emergency of international concern in July 2022, and prophylactic and therapeutic measures are in urgent need. The monkeypox virus (MPXV) has its own DNA polymerase F8, together with the processive cofactors A22 and E4, constituting the polymerase holoenzyme for genome replication. Here, we determined the holoenzyme structure in complex with DNA using cryo-electron microscopy at the global resolution of ~2.8 angstroms. The holoenzyme possesses an architecture that suggests a "forward sliding clamp" processivity mechanism for viral DNA replication. MPXV polymerase has a DNA binding mode similar to that of other B-family DNA polymerases from different species. These findings reveal the mechanism of the MPXV genome replication and may guide the development of anti-poxvirus drugs.
Organizational Affiliation:
CAS Key Laboratory of Pathogen Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China.