Exploring AlphaFold2's Performance on Predicting Amino Acid Side-Chain Conformations and Its Utility in Crystal Structure Determination of B318L Protein.
Zhao, H., Zhang, H., She, Z., Gao, Z., Wang, Q., Geng, Z., Dong, Y.(2023) Int J Mol Sci 24
- PubMed: 36769074
- DOI: https://doi.org/10.3390/ijms24032740
- Primary Citation of Related Structures:
8HDL - PubMed Abstract:
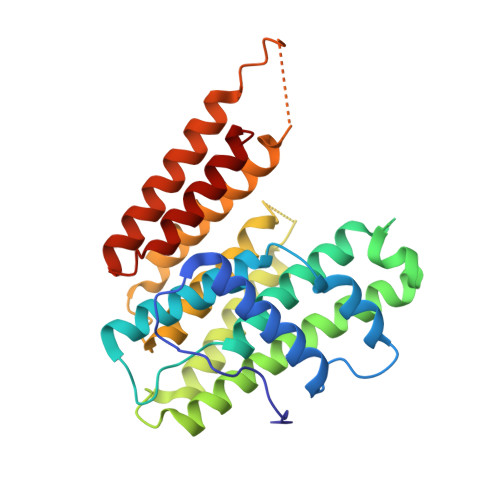
Recent technological breakthroughs in machine-learning-based AlphaFold2 (AF2) are pushing the prediction accuracy of protein structures to an unprecedented level that is on par with experimental structural quality. Despite its outstanding structural modeling capability, further experimental validations and performance assessments of AF2 predictions are still required, thus necessitating the development of integrative structural biology in synergy with both computational and experimental methods. Focusing on the B318L protein that plays an essential role in the African swine fever virus (ASFV) for viral replication, we experimentally demonstrate the high quality of the AF2 predicted model and its practical utility in crystal structural determination. Structural alignment implies that the AF2 model shares nearly the same atomic arrangement as the B318L crystal structure except for some flexible and disordered regions. More importantly, side-chain-based analysis at the individual residue level reveals that AF2's performance is likely dependent on the specific amino acid type and that hydrophobic residues tend to be more accurately predicted by AF2 than hydrophilic residues. Quantitative per-residue RMSD comparisons and further molecular replacement trials suggest that AF2 has a large potential to outperform other computational modeling methods in terms of structural determination. Additionally, it is numerically confirmed that the AF2 model is accurate enough so that it may well potentially withstand experimental data quality to a large extent for structural determination. Finally, an overall structural analysis and molecular docking simulation of the B318L protein are performed. Taken together, our study not only provides new insights into AF2's performance in predicting side-chain conformations but also sheds light upon the significance of AF2 in promoting crystal structural determination, especially when the experimental data quality of the protein crystal is poor.
Organizational Affiliation:
School of Life Sciences, University of Science and Technology of China, Hefei 230027, China.