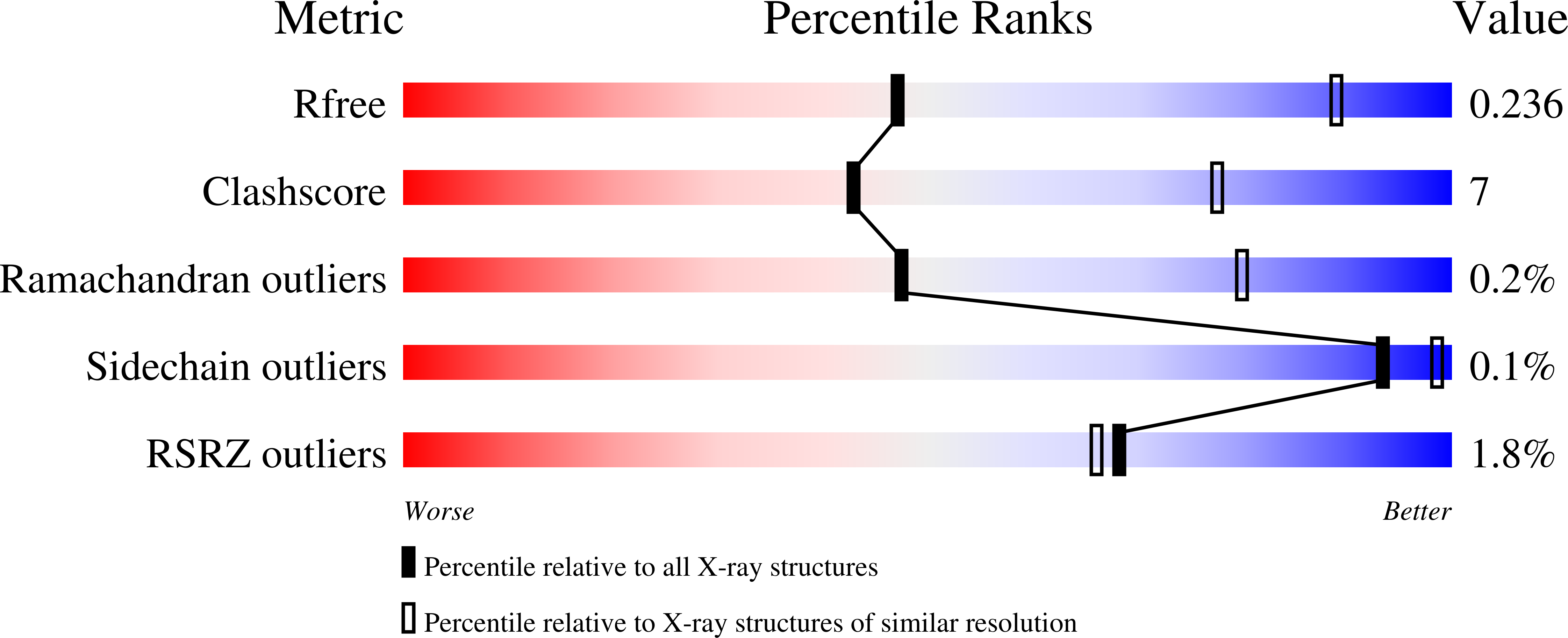
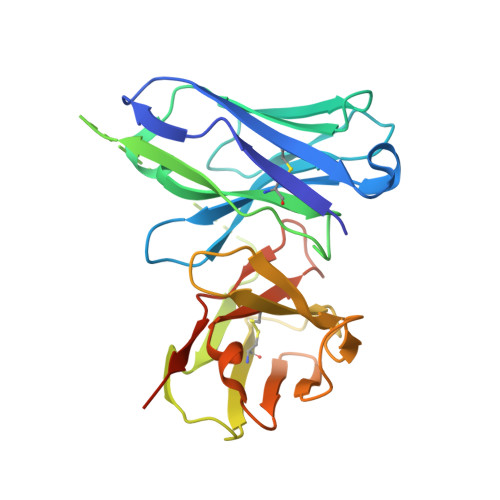
A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
Kumar, U., Goyal, P., Madni, Z.K., Kamble, K., Gaur, V., Rajala, M.S., Salunke, D.M.(2023) J Biomed Sci 30: 56-56
- PubMed: 37491224
- DOI: https://doi.org/10.1186/s12929-023-00950-2
- Primary Citation of Related Structures:
8H3B, 8H3C, 8H73 - PubMed Abstract:
The influenza virus enters the host via hemagglutinin protein binding to cell surface sialic acid. Receptor-mediated endocytosis is followed by viral nucleocapsid uncoating for replication aided by the transmembrane viral M2 proton ion channel. M2 ectodomain (M2e) is a potential universal candidate for monoclonal antibody therapy owing to its conserved nature across influenza virus subtypes and its importance in viral propagation.
Organizational Affiliation:
International Centre for Genetic Engineering and Biotechnology, Aruna Asaf Ali Marg, New Delhi, 110067, India.