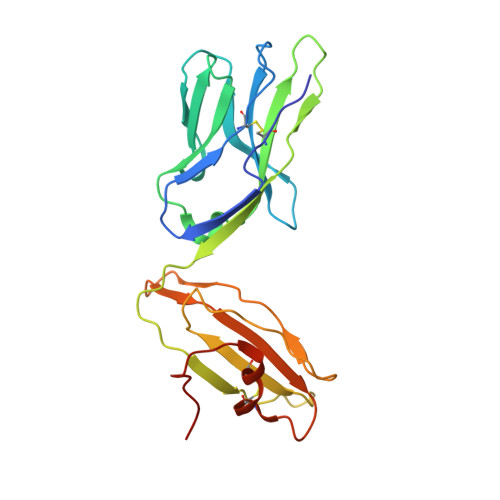
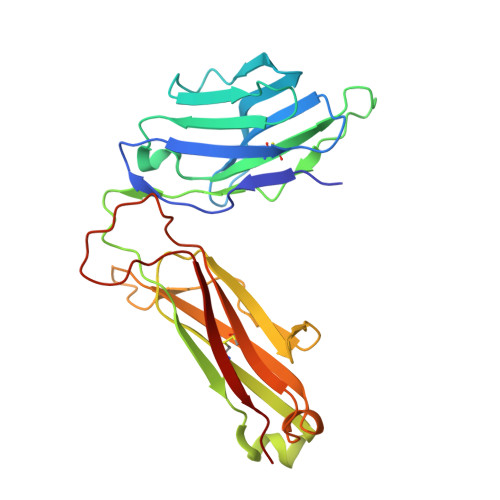
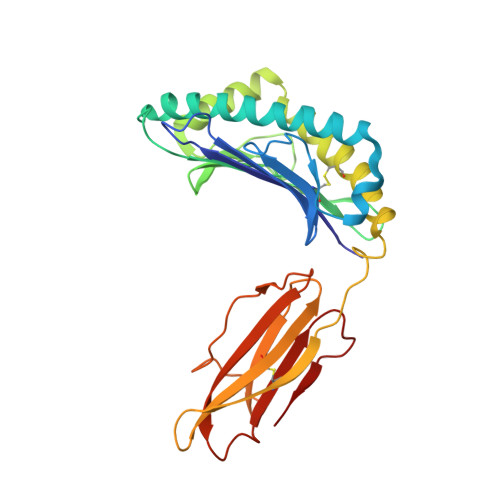
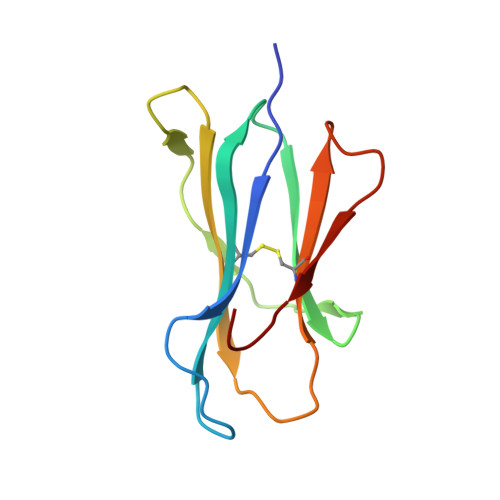
Molecular Basis for the Recognition of HIV Nef138-8 Epitope by a Pair of Human Public T Cell Receptors.
Ma, K., Chai, Y., Guan, J., Tan, S., Qi, J., Kawana-Tachikawa, A., Dong, T., Iwamoto, A., Shi, Y., Gao, G.F.(2022) J Immunol 209: 1652-1661
- PubMed: 36130828
- DOI: https://doi.org/10.4049/jimmunol.2200191
- Primary Citation of Related Structures:
8GVB, 8GVG, 8GVI - PubMed Abstract:
Cross-recognized public TCRs against HIV epitopes have been proposed to be important for the control of AIDS disease progression and HIV variants. The overlapping Nef138-8 and Nef138-10 peptides from the HIV Nef protein are HLA-A24-restricted immunodominant T cell epitopes, and an HIV mutant strain with a Y139F substitution in Nef protein can result in immune escape and is widespread in Japan. Here, we identified a pair of public TCRs specific to the HLA-A24-restricted Nef-138-8 epitope using PBMCs from White and Japanese patients, respectively, namely TD08 and H25-11. The gene use of the variable domain for TD08 and H25-11 is TRAV8-3, TRAJ10 for the α-chain and TRBV7-9, TRBD1*01, TRBJ2-5 for the β-chain. Both TCRs can recognize wild-type and Y2F-mutated Nef138-8 epitopes. We further determined three complex structures, including TD08/HLA-A24-Nef138-8, H25-11/HLA-A24-Nef138-8, and TD08/HLA-A24-Nef138-8 (2F). Then, we revealed the molecular basis of the public TCR binding to the peptide HLA, which mostly relies on the interaction between the TCR and HLA and can tolerate the mutation in the Nef138-8 peptide. These findings promote the molecular understanding of T cell immunity against HIV epitopes and provide an important basis for the engineering of TCRs to develop T cell-based immunotherapy against HIV infection.
Organizational Affiliation:
Savaid Medical School, University of Chinese Academy of Sciences, Beijing, China.